
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,497,490 – 20,497,610 |
| Length | 120 |
| Max. P | 0.628292 |

| Location | 20,497,490 – 20,497,610 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.20 |
| Shannon entropy | 0.70589 |
| G+C content | 0.52500 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -15.64 |
| Energy contribution | -14.21 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.79 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.628292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20497490 120 - 24543557 ACAUCAUGAAUGGAAUUGGAGCGUAGGUGGUUGCCACUGCCAGUAGCAUCACUACUUUCCGGCCAUGAAAAUCGGAAAUGGCACCCAUCACGGGCGUGGUCACAAAUCCCAGGAUUCCUC ...........((((((((((.((((.((((.((.((.....)).)))))))))).))))(((((((....(((...((((...))))..))).)))))))...........)))))).. ( -36.20, z-score = -0.32, R) >droSim1.chr3L 19847320 120 - 22553184 ACAUCAUGAAUGGAAUUGGAGCGUAGGUGGUGGCCACAGCCAGUAGCAUCACCACUUUUCGGCCACGGAAAUCGGAAAUGGCGCCCAUCACAGGCGUGGUCACAAAUCCCAGGAUUCCUC ...........(((((((((((....)).((((((((.(((((......).((..((((((....))))))..))...))))(((.......)))))))))))...)))...)))))).. ( -37.30, z-score = -0.38, R) >droSec1.super_11 387871 120 - 2888827 ACAUCAUGAAUGGAAUUGGAGCGUAGGUGGUGGCCACAGCCAGUAGCAUCACCACUUUUCGGCCACGGAAAUCGGAAAUGGCGCCCAUCACAGGCGUGGUCACAAAUCCCAGGAUUCCUC ...........(((((((((((....)).((((((((.(((((......).((..((((((....))))))..))...))))(((.......)))))))))))...)))...)))))).. ( -37.30, z-score = -0.38, R) >droYak2.chr3L 4137941 120 + 24197627 ACAUCAUGAAUGGAAUCGGCGCGUAAGUGGUUGCCACGGCCAGCAACAUCACCACUUUUCGGCCACGGAAAUCAGAAAUGGCGCCCAUCACAGGUGUAGUCACAAAUCCCAGGAUUCCUC ...........(((((((((.((.((((((((((........))).....)))))))..)))))..(((...((....))(((((.......))))).........)))...)))))).. ( -34.90, z-score = -0.92, R) >droEre2.scaffold_4784 20189336 120 - 25762168 ACAUCAUGAAUGGAAUUGGCGCGUAAGUGGUGGCCACAGCUAGCAACAUCACCACUUUUCGGCCACGGAAAUCAGAAAUGGCGCCCAUUACAGGUGUGGUCAGAAAUCCCAGGAUUCCUC ...........(((((((((.((.((((((((((........)).....))))))))..)))))..(((..((....((.(((((.......))))).))..))..)))...)))))).. ( -33.50, z-score = -0.08, R) >droAna3.scaffold_13337 10851167 120 - 23293914 UGACCAUGAAGGGGAUGGGCGAGUAGGUGGUGGCCACUGCCAGUAGCAUCACGAUUUUCCUGCCCCGGAAGUCGGAUAGGGCGCCCAGUAGAGGCGUGGUCAGGAAGGAGACCAUUCCCC ((((((((.......((((((....(.((.((((....)))).)).)(((.((((((.((......)))))))))))....)))))).......))))))))(((((....)..)))).. ( -46.54, z-score = -0.99, R) >dp4.chrXR_group8 7901873 120 - 9212921 UGAUCAUCAUCGGAAUGGGCGAGUAGGUGGUCACCACGGCUAUCAGCAUCAGCGGCUUGCGUCCCCAAAUGUCCGAGAGGGCGCCCAUCAGCGGGGCCGCGAUGAAGGCAAGCGUUCCCC ...........((((((.((((((.(((((((.....))))))).((....)).))))))..........((((....))))(((((((.(((....)))))))..)))...)))))).. ( -45.20, z-score = -0.17, R) >droPer1.super_40 91620 120 - 810552 UGAUCAUCAUCGGAAUGGGCGAGUAGGUGGUCACCACGGCUAUCAGCAUCAGCGGCUUGCGUCCCCAAAUGUCCGAGAGGGCGCCCAUCAGCGGGACCGCGAUGAAGGCAAACGUUCCCC ...........((((((.((((((.(((((((.....))))))).((....)).))))))..........((((....))))(((((((.(((....)))))))..)))...)))))).. ( -45.50, z-score = -0.93, R) >droWil1.scaffold_181009 3027140 120 - 3585778 UGGACAUCAACGGUAUGGGUAGGCAGGUAAAGAAUACUGUGACUAAUAGGAAAAAUUUGCGACCCCAGAUAUCAGAGAGGGCGCCAAUUAAUGGCGCCGACAGGAAUGACAAUAUGCCCU .((.(((....(((((((((..((((((........((((.....)))).....)))))).))))...)))))......(((((((.....)))))))...............))).)). ( -32.02, z-score = -1.33, R) >droVir3.scaffold_13049 25029165 120 + 25233164 CGCACAUCAGCGGUAUCGGCAUACAUGUAAAGAAUACUGUAACUAAUAGGAAAAAUUUACGACCCCAGAUGUCGGACAGGGCGCCUAUCAAUGGCGCCGACAGGAAUGACAAUAUGCCCU (((......))).....((((((..((((((.....((((.....))))......))))))........(((((..(..((((((.......)))))).....)..))))).)))))).. ( -33.40, z-score = -1.72, R) >droMoj3.scaffold_6680 952859 120 - 24764193 UGCACAUCAGCGGUAUGGGCAUACAGGUGAAGAAUACUGUAACUAAUAGGAAAAACUUGCGACCCCAGAUGUCCGACAGAGCGCCGAUCAAGGGGGCCGACAGAAAGGACAGUAUGCCCU .((......)).....((((((((.(((((((....((((.....))))......))).).))).....(((((....(.((.((.......)).)))........))))))))))))). ( -31.50, z-score = -0.20, R) >droGri2.scaffold_15110 23041327 120 + 24565398 UGCACAUCAGCGGUAUGGGCAUACAUGUGAAGAAUACUGUUACUAAUAGGAAGAAUUUACGUCCCCAGAUGUCGGAGAGGGCGCCAAUCAGUGGCGCCGACAGGAAUGACAGUAUGCCCU .((......)).....((((((((..(((((.....((((.....))))......)))))(((......((((......(((((((.....))))))))))).....))).)))))))). ( -39.90, z-score = -2.16, R) >apiMel3.Group15 6953885 120 - 7856270 UGCUCAUUAGAGGAAUCGGCGCACAUGUGAAGGCUACUGUGAUGAGUAAAAAGAAUUUUCGACCCCAUACAUCAGAUAAAGCACCAAUUAAUGGUGCAGAAAGAAAUGAUAAGAUUCCUU (((((((((.((...((..(((....)))..))...)).)))))))))....(((((((...........((((......((((((.....)))))).........)))))))))))... ( -28.36, z-score = -1.79, R) >consensus UGAUCAUCAACGGAAUGGGCGCGUAGGUGGUGGCCACUGCCAGUAGCAUCACCAAUUUUCGGCCCCAGAAAUCGGAAAGGGCGCCCAUCACAGGCGCCGUCAGAAAUGCCAGGAUUCCCC ...........((((((.........(((.....)))...........................................(((((.......)))))...............)))))).. (-15.64 = -14.21 + -1.44)

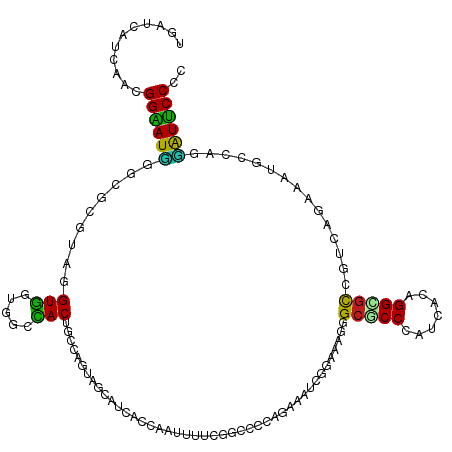

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:03 2011