
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,489,699 – 20,489,819 |
| Length | 120 |
| Max. P | 0.514468 |

| Location | 20,489,699 – 20,489,819 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.90 |
| Shannon entropy | 0.43426 |
| G+C content | 0.55922 |
| Mean single sequence MFE | -43.84 |
| Consensus MFE | -22.94 |
| Energy contribution | -22.50 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
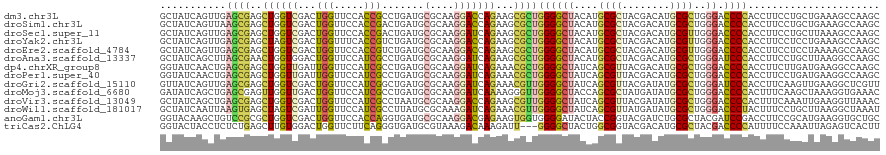
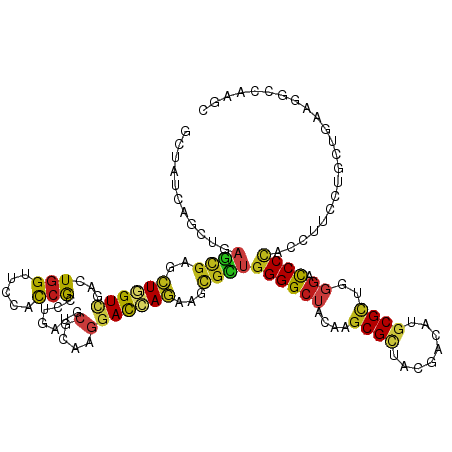
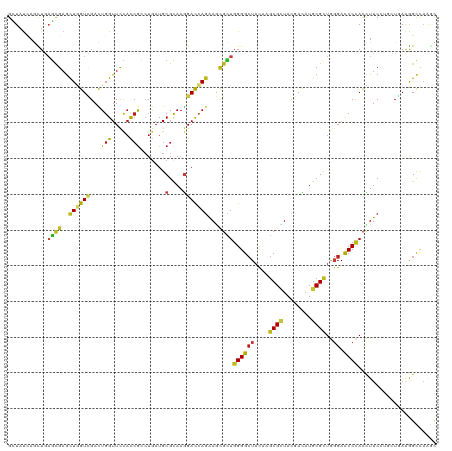
>dm3.chr3L 20489699 120 + 24543557 GCUAUCAGUUGAGCGAGCUGGUCGACUGGUUCCACCGCCUGAUGCGCAAGGACCAGAAGCGCUGGGGCUACAUGCGCUACGACAUGCGCUGGGACCCCACCUUCCUGCUGAAAGCCAAGC (((.(((((.(((((..((((((....(((......))).....(....)))))))...))))((((((.((.((((........)))))))).))))......).))))).)))..... ( -46.80, z-score = -1.35, R) >droSim1.chr3L 19839339 120 + 22553184 GCUAUCAGUUAAGCGAGCUGGUCGACUGGUUCCACCGACUGAUGCGCAAGGACCAGAAGCGCUGGGGCUACAUGCGCUACGACAUGCGCUGGGACCCCACCUUCCUGCUGAAAGCCAAGC (((.(((((..((((..((((((...(((.....))).......(....)))))))...))))((((((.((.((((........)))))))).))))........))))).)))..... ( -45.90, z-score = -1.76, R) >droSec1.super_11 380278 120 + 2888827 GCUAUCAGUUGAGCGAGCUGGUCGACUGGUUCCACCGACUGAUGCGCAAGGAUCAGAAGCGCUGGGGCUACAUGCGCUACGACAUGCGUUGGGACCCCACCUUCCUGCUUAAAGCCAAGC (((.....(((((((.(..(((.(...(((((((.((.(((((.(....).))))).(((((((......)).)))))........)).)))))))).)))..).))))))).....))) ( -43.40, z-score = -1.05, R) >droYak2.chr3L 4128219 120 - 24197627 GCUAUCAGUUGAGCGAGCUAGUCGACUGGUUUCACCGUCUGAUGCGCAAGGACCAGAAGCGCUGGGGCUACAUGCGCUACGACAUGCGUUGGGACCCCACCUUCCUCCUGAAAGCCAAGC (((.((((..(((.((((..((((.(((((((...(((.....)))...))))))).(((((((......)).))))).))))..))(.(((....))).).))))))))).)))..... ( -40.60, z-score = -0.46, R) >droEre2.scaffold_4784 20182828 120 + 25762168 GCUAUCAGUUGAGCGAGCUGGUCGACUGGUUCCACCGUCUGAUGCGCAAGGACCAGAAGCGCUGGGGCUACAUGCGCUACGACAUGCGUUGGGACCCCACCUUCCUCCUAAAAGCCAAGC (((........((((..(((((((((.((.....))))).....(....)))))))...))))((((((....((((........))))..)).))))..............)))..... ( -40.60, z-score = -0.30, R) >droAna3.scaffold_13337 10847753 120 + 23293914 GCUAUCAGCUUAGCGAACUGGUGGACUGGUUCCAUCGCCUGAUGCGCAAGGAUCAGAAGCGCUGGGGCUACAUGCGCUACGACAUGCGCUGGGAUCCCACCUUCCUGCUUAAGGCCAAGC (((....((((((((....(((((..((((((((.((((((((.(....).)))))..))).))))))))((.((((........)))))).....)))))....))))..))))..))) ( -47.50, z-score = -1.66, R) >dp4.chrXR_group8 7898233 120 + 9212921 GGUAUCAACUGAGCGAGCUGGUUGAUUGGUUCCAUCGCCUGAUGCGCAAGGAUCAGAAACGCUGGGGCUAUCAGCGUUACGACAUGCGCUGGGACCCCACCUUCUUGAUGAAGGCCAAGC ((.(((((((.((....))))))))).(((((((.((((((((.(....).))))).((((((((.....)))))))).......))).))))))))).(((((.....)))))...... ( -49.50, z-score = -2.95, R) >droPer1.super_40 87906 120 + 810552 GGUAUCAACUGAGCGAGCUGGUUGAUUGGUUCCAUCGCCUGAUGCGCAAGGAUCAGAAACGCUGGGGCUAUCAGCGUUACGACAUGCGCUGGGACCCCACCUUCCUGAUGAAGGCCAAGC ((.(((((((.((....))))))))).(((((((.((((((((.(....).))))).((((((((.....)))))))).......))).))))))))).(((((.....)))))...... ( -48.30, z-score = -2.49, R) >droGri2.scaffold_15110 4745964 120 - 24565398 GUUAUCAGUUGAGCGAGCUGGUCGACUGGUUCCAUCGGCUGAUGCGCAAGGAUCAGAAACGUUGGGGCUAUCAGCGUUACGAUAUGCGCUGGGAUCCCACCUUCAAGUUGAAGGCUCGUU ...........((((((((..(((((((((((((.((.(((((.(....).)))))...)).))))))).(((((((........))))))).............)))))).)))))))) ( -50.20, z-score = -3.32, R) >droMoj3.scaffold_6680 12371535 120 + 24764193 GAUAUCAGCUGAGCGAGUUGGUUGACUGGUUCCAUCGCCUGAUGCGCAAGGAUCAAAAGGGUUGGGGCUACCAGCGCUAUGAUAUGCGCUGGGACCCCACUUUCAAGCUAAAGGUGAAAC ..........((((.((((....)))).))))..(((((((((.(....).)))...(((((.((((...(((((((........)))))))..)))))))))........))))))... ( -47.40, z-score = -3.12, R) >droVir3.scaffold_13049 6379801 120 - 25233164 GCUAUCAGCUGAGCGAGCUGGUCGACUGGUUCCAUCGCCUAAUGCGCAAGGACCAGAAGCGUUGGGGCUAUCAGCGUUACGAUAUGCGCUGGGACCCCACUUUCAAAUUGAAGGUUAAAC ((((((((((.....)))))))...(((((((...(((.....)))...))))))).)))...((((...(((((((........)))))))..))))((((((.....))))))..... ( -46.90, z-score = -2.96, R) >droWil1.scaffold_181017 475870 120 - 939937 GCUAUCAAUUAAGUGAGCUAGUCGAUUGGUUCCAUCGCCUUAUGCGCAAAGAUCAGAAACGUUGGGGCUAUCAGCGUUAUGAUAUGCGCUGGGACCCUACUUUCCUGCUUAAGGCUAAAU ............(.(((((((....))))))))...((((((.(((((...((((..((((((((.....)))))))).)))).))))).((((........))))...))))))..... ( -36.40, z-score = -1.40, R) >anoGam1.chr3L 9034459 120 + 41284009 GGUACAAGCUGUCCGCGCUGGUCGACUGGUUCCACCAGGUGAUGCGCAAGGACGAGAAGUGGUGGGGAUACUACCGGUACGAUCUGCGCUACGAUCCGACCUUCCGCAUGAAGGUGCUGC ((.((.....))))(((((..((..((((.....))))...(((((.((((.((.((.((((((.((((..((....))..)))).))))))..)))).)))).))))))).)))))... ( -43.20, z-score = -0.15, R) >triCas2.ChLG4 3851965 117 + 13894384 GGUACUACCUCUCUGAGCUUGUGGACUGGUUCUUCAGGGUGAUGCGUAAAGACAAAGAUU---GGGGCUACUGGCGGUACGACAUGCGCUACGACCCCAUUUUCCAAAUUAGAGUCACUU .(((.(((((....(((((........)))))....))))).))).....(((......(---((((....(((((..........)))))...)))))...((.......))))).... ( -27.10, z-score = 1.50, R) >consensus GCUAUCAGCUGAGCGAGCUGGUCGACUGGUUCCACCGCCUGAUGCGCAAGGACCAGAAGCGCUGGGGCUACAAGCGCUACGACAUGCGCUGGGACCCCACCUUCCUGCUGAAGGCCAAGC ...........((((..((((((...(((.....))).......(....)))))))...))))((((((....((((........))))..)).))))...................... (-22.94 = -22.50 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:00 2011