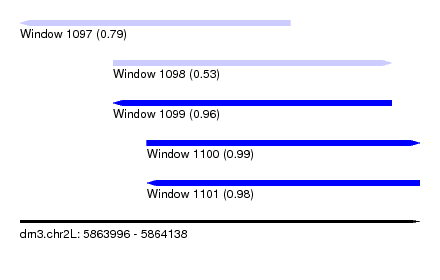
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,863,996 – 5,864,138 |
| Length | 142 |
| Max. P | 0.994862 |

| Location | 5,863,996 – 5,864,092 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.41 |
| Shannon entropy | 0.29177 |
| G+C content | 0.38284 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -12.93 |
| Energy contribution | -13.60 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.789277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5863996 96 - 23011544 UGCCGGUUAGAUAUGGAUACGUGAGUAUCCACUUUCACCGAUUGCUUUCUGCUUAAAGCCCAAAAAACACGAUCCAU---AUAUGA----AUGCAAUAUCAUU (((((((.(((..(((((((....))))))).))).))))...(((((......)))))..................---......----..)))........ ( -22.20, z-score = -2.41, R) >droYak2.chr2L 8990529 91 + 22324452 UUCCAGUUAGAUAUGGAUACGUGAGUCGCCAC-----------AAUUUCUGCUUUAAA-CCCAACAUCACGAUCCAUAUGAUAUGAUGAUGUGCAAUAUCAUU ..........((((((((.(((((........-----------...............-.......))))))))))))).....((((((((...)))))))) ( -18.01, z-score = -0.86, R) >droSim1.chr2L 5654530 95 - 22036055 UGCCGGUUAGAUAUGGAUACGUGAGUAUCCACUUUCACCGAUUGCUUUCUGCUUUAAG-CAGAACAACACGAUCCAU---AUAUGA----AUGCAAUAUCAUU (((.(((.(((..(((((((....))))))).))).)))(((((..((((((.....)-))))).....)))))...---......----..)))........ ( -27.60, z-score = -3.84, R) >consensus UGCCGGUUAGAUAUGGAUACGUGAGUAUCCACUUUCACCGAUUGCUUUCUGCUUUAAG_CCAAACAACACGAUCCAU___AUAUGA____AUGCAAUAUCAUU .........(((((((((((....))))))...................(((........................................))))))))... (-12.93 = -13.60 + 0.67)



| Location | 5,864,029 – 5,864,128 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 66.40 |
| Shannon entropy | 0.58660 |
| G+C content | 0.43834 |
| Mean single sequence MFE | -23.34 |
| Consensus MFE | -9.42 |
| Energy contribution | -10.50 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.532141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5864029 99 + 23011544 UUGGGCUUUAAGCAGAAAGCAAUCGGUGAAAGUGGAUACUC-ACGUAUCCAUAU-CUAACCGGCACUUCUAGCAGGGCCUU-UCCGCAUCAAGAGUCAUAAA ...((((((..((.((((((...((((....((((((((..-..))))))))..-...))))((.......))...).)))-)).))....))))))..... ( -28.40, z-score = -2.15, R) >droAna3.scaffold_12916 11275690 99 + 16180835 -UCCGAUUAAAACGAGGUAUAAGCG--AAUCGUAACCUCUCGAAAGAAGCAUACAUUGUCUUGCAUGCACCGCUGAGAUUUAUCAGCUUAAAGGGCAAUAAA -(((.........(((((....(((--...))).)))))((....)).((((.((......)).))))...(((((......))))).....)))....... ( -22.10, z-score = -1.11, R) >droYak2.chr2L 8990569 87 - 22324452 ------------UGGGUUUAAAGCAGAAAUUGUGGCGACUC-ACGUAUCCAUAU-CUAACUGGAACUUCUAGCAAGGCUUU-UCCGCGUCAAGAGUCAUAAA ------------..........((((((...((((....))-))((.((((...-.....)))))))))).))..((((((-(.......)))))))..... ( -15.20, z-score = 0.87, R) >droSec1.super_5 3933892 85 + 5866729 --------------GAAAGCAAUCGGUGAAAGUGGAUACUC-ACGUAUCCAUAU-CUAACCGGCACUUCUAGCAGGGCUUU-UCCGCGUCAAGAGUCAUAAA --------------..........(((....((((((((..-..))))))))..-...)))(((.(((...((.(((....-)))))...))).)))..... ( -22.30, z-score = -1.80, R) >droSim1.chr2L 5654563 98 + 22036055 -UCUGCUUAAAGCAGAAAGCAAUCGGUGAAAGUGGAUACUC-ACGUAUCCAUAU-CUAACCGGCACUUCUAGCAGGGCUUU-UCCGCGUCAAGAGUCAUAAA -(((((.....)))))........(((....((((((((..-..))))))))..-...)))(((.(((...((.(((....-)))))...))).)))..... ( -28.70, z-score = -2.67, R) >consensus _U__G_UU_AA_CAGAAAGCAAUCGGUGAAAGUGGAUACUC_ACGUAUCCAUAU_CUAACCGGCACUUCUAGCAGGGCUUU_UCCGCGUCAAGAGUCAUAAA ...............................((((((((.....)))))))).........(((.(((...((.((......)).))...))).)))..... ( -9.42 = -10.50 + 1.08)

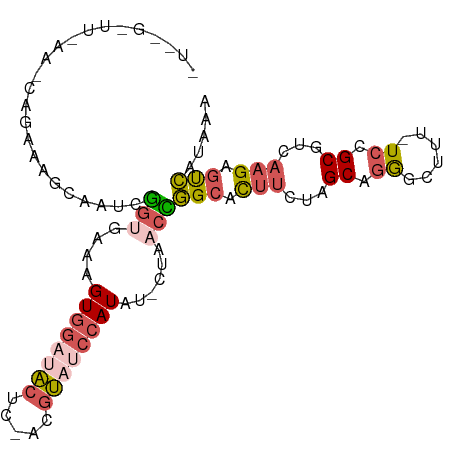

| Location | 5,864,029 – 5,864,128 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 66.40 |
| Shannon entropy | 0.58660 |
| G+C content | 0.43834 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -10.62 |
| Energy contribution | -11.02 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.962130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5864029 99 - 23011544 UUUAUGACUCUUGAUGCGGA-AAGGCCCUGCUAGAAGUGCCGGUUAG-AUAUGGAUACGU-GAGUAUCCACUUUCACCGAUUGCUUUCUGCUUAAAGCCCAA .........(((...(((((-(((((.((......)).))((((.((-(..(((((((..-..))))))).))).))))....))))))))...)))..... ( -32.00, z-score = -3.22, R) >droAna3.scaffold_12916 11275690 99 - 16180835 UUUAUUGCCCUUUAAGCUGAUAAAUCUCAGCGGUGCAUGCAAGACAAUGUAUGCUUCUUUCGAGAGGUUACGAUU--CGCUUAUACCUCGUUUUAAUCGGA- .......((..(((((((((......)))))((.(((((((......))))))).))......(((((.......--.......)))))...))))..)).- ( -23.74, z-score = -1.10, R) >droYak2.chr2L 8990569 87 + 22324452 UUUAUGACUCUUGACGCGGA-AAAGCCUUGCUAGAAGUUCCAGUUAG-AUAUGGAUACGU-GAGUCGCCACAAUUUCUGCUUUAAACCCA------------ ..........((((.(((((-((...((.(((.(.....).))).))-...(((..((..-..))..)))...))))))).)))).....------------ ( -17.50, z-score = -0.39, R) >droSec1.super_5 3933892 85 - 5866729 UUUAUGACUCUUGACGCGGA-AAAGCCCUGCUAGAAGUGCCGGUUAG-AUAUGGAUACGU-GAGUAUCCACUUUCACCGAUUGCUUUC-------------- ...............((((.-......))))..(((((..((((.((-(..(((((((..-..))))))).))).))))...))))).-------------- ( -23.80, z-score = -2.13, R) >droSim1.chr2L 5654563 98 - 22036055 UUUAUGACUCUUGACGCGGA-AAAGCCCUGCUAGAAGUGCCGGUUAG-AUAUGGAUACGU-GAGUAUCCACUUUCACCGAUUGCUUUCUGCUUUAAGCAGA- .......(((((((.(((((-((.((...((.......))((((.((-(..(((((((..-..))))))).))).))))...))))))))).))))).)).- ( -33.30, z-score = -3.81, R) >consensus UUUAUGACUCUUGACGCGGA_AAAGCCCUGCUAGAAGUGCCGGUUAG_AUAUGGAUACGU_GAGUAUCCACUUUCACCGAUUGCUUUCCG_UU_AA_C__A_ ...............((((........)))).........((((.......(((((((.....))))))).....))))....................... (-10.62 = -11.02 + 0.40)

| Location | 5,864,041 – 5,864,138 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 68.57 |
| Shannon entropy | 0.54786 |
| G+C content | 0.46568 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -15.62 |
| Energy contribution | -17.82 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.994862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5864041 97 + 23011544 -----CAGAAAGCAAUCGGUGAAAGUGGAUACUCACGUAUCCAUAUCUAACCGGCACUUCUAGCAGGGCCUUUCCGCAUCAAGAGUCAUAAAGCCUGCUACC-- -----.((((.((...((((....((((((((....)))))))).....))))))..))))((((((..((((..((.......))...))))))))))...-- ( -26.30, z-score = -1.96, R) >droAna3.scaffold_12916 11275701 100 + 16180835 CGAGGUAUAAGCGAAUCGUAACCUCUCGAAAGAAGCAUA-CAUUGUCUUGCAUGCACCGCUG---AGAUUUAUCAGCUUAAAGGGCAAUAAAAUUUUUUCAGCC .(((((....(((...))).)))))((....)).((((.-((......)).))))...((((---((((((((..(((.....))).)))))....))))))). ( -23.80, z-score = -1.37, R) >droYak2.chr2L 8990580 86 - 22324452 ----------------CAGAAAUUGUGGCGACUCACGUAUCCAUAUCUAACUGGAACUUCUAGCAAGGCUUUUCCGCGUCAAGAGUCAUAAAGCCUGCUUCC-- ----------------.((((...((((....))))((.((((........))))))))))((((.((((((...((.......))...))))))))))...-- ( -19.50, z-score = -0.86, R) >droSec1.super_5 3933892 95 + 5866729 -------GAAAGCAAUCGGUGAAAGUGGAUACUCACGUAUCCAUAUCUAACCGGCACUUCUAGCAGGGCUUUUCCGCGUCAAGAGUCAUAAAGCCUGCUACC-- -------....((...((((....((((((((....)))))))).....)))))).....(((((((.((((...((.......))...)))))))))))..-- ( -30.40, z-score = -3.24, R) >droSim1.chr2L 5654574 97 + 22036055 -----CAGAAAGCAAUCGGUGAAAGUGGAUACUCACGUAUCCAUAUCUAACCGGCACUUCUAGCAGGGCUUUUCCGCGUCAAGAGUCAUAAAGCCUGCUACC-- -----.((((.((...((((....((((((((....)))))))).....))))))..))))((((((.((((...((.......))...))))))))))...-- ( -30.40, z-score = -3.07, R) >consensus ______AGAAAGCAAUCGGUGAAAGUGGAUACUCACGUAUCCAUAUCUAACCGGCACUUCUAGCAGGGCUUUUCCGCGUCAAGAGUCAUAAAGCCUGCUACC__ ................((((....((((((((....)))))))).....)))).......((((.(((((((...((.......))...))))))))))).... (-15.62 = -17.82 + 2.20)

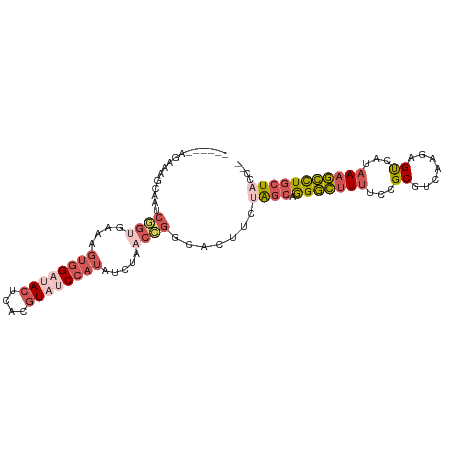

| Location | 5,864,041 – 5,864,138 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 68.57 |
| Shannon entropy | 0.54786 |
| G+C content | 0.46568 |
| Mean single sequence MFE | -28.39 |
| Consensus MFE | -15.45 |
| Energy contribution | -15.37 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.975906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5864041 97 - 23011544 --GGUAGCAGGCUUUAUGACUCUUGAUGCGGAAAGGCCCUGCUAGAAGUGCCGGUUAGAUAUGGAUACGUGAGUAUCCACUUUCACCGAUUGCUUUCUG----- --..(((((((((((.((.(.......)))..)))).)))))))((((.((((((.(((..(((((((....))))))).))).))))...))))))..----- ( -32.50, z-score = -2.34, R) >droAna3.scaffold_12916 11275701 100 - 16180835 GGCUGAAAAAAUUUUAUUGCCCUUUAAGCUGAUAAAUCU---CAGCGGUGCAUGCAAGACAAUG-UAUGCUUCUUUCGAGAGGUUACGAUUCGCUUAUACCUCG (((..(((....)))...)))......(((((......)---))))((.(((((((......))-))))).))......(((((..............))))). ( -24.44, z-score = -1.37, R) >droYak2.chr2L 8990580 86 + 22324452 --GGAAGCAGGCUUUAUGACUCUUGACGCGGAAAAGCCUUGCUAGAAGUUCCAGUUAGAUAUGGAUACGUGAGUCGCCACAAUUUCUG---------------- --...((((((((((.....(((......))))))))).))))((((((((((........))))...(((......))).)))))).---------------- ( -20.30, z-score = -0.06, R) >droSec1.super_5 3933892 95 - 5866729 --GGUAGCAGGCUUUAUGACUCUUGACGCGGAAAAGCCCUGCUAGAAGUGCCGGUUAGAUAUGGAUACGUGAGUAUCCACUUUCACCGAUUGCUUUC------- --..(((((((((((.....(((......))))))).)))))))(((((..((((.(((..(((((((....))))))).))).))))...))))).------- ( -32.30, z-score = -2.80, R) >droSim1.chr2L 5654574 97 - 22036055 --GGUAGCAGGCUUUAUGACUCUUGACGCGGAAAAGCCCUGCUAGAAGUGCCGGUUAGAUAUGGAUACGUGAGUAUCCACUUUCACCGAUUGCUUUCUG----- --..(((((((((((.....(((......))))))).)))))))((((.((((((.(((..(((((((....))))))).))).))))...))))))..----- ( -32.40, z-score = -2.63, R) >consensus __GGUAGCAGGCUUUAUGACUCUUGACGCGGAAAAGCCCUGCUAGAAGUGCCGGUUAGAUAUGGAUACGUGAGUAUCCACUUUCACCGAUUGCUUUCU______ ..(((((((((((((.................))))).)))))..................(((((((....))))))).....)))................. (-15.45 = -15.37 + -0.08)

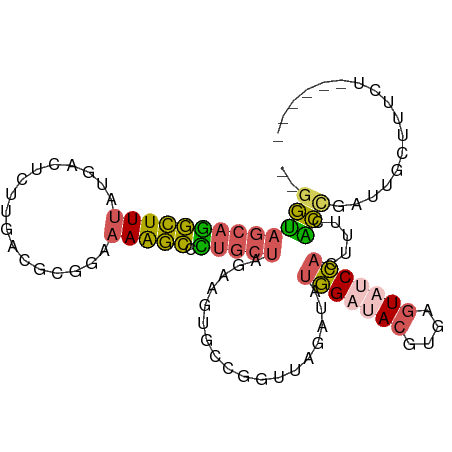
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:59 2011