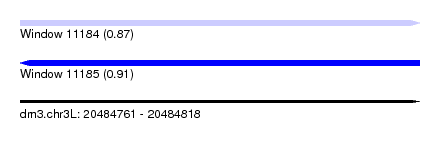
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,484,761 – 20,484,818 |
| Length | 57 |
| Max. P | 0.912846 |

| Location | 20,484,761 – 20,484,818 |
|---|---|
| Length | 57 |
| Sequences | 6 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 70.27 |
| Shannon entropy | 0.58165 |
| G+C content | 0.44664 |
| Mean single sequence MFE | -13.85 |
| Consensus MFE | -6.79 |
| Energy contribution | -7.54 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.873591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
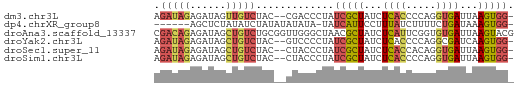
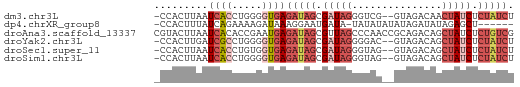
>dm3.chr3L 20484761 57 + 24543557 AGAUAGAGAUAGUUGUCUAC--CGACCCUAUCGCUAUCUCACCCCAGGUGAUUAAGUGG- .(((((.(((((..(((...--.))).))))).)))))(((((...)))))........- ( -18.00, z-score = -3.00, R) >dp4.chrXR_group8 7892956 52 + 9212921 ------AGCUCUAUAUCUAUAUAUAUA-UAUCAUUCCUUUAUCUUUUCUGAUAAAGUGG- ------.....................-........(((((((......)))))))...- ( -6.20, z-score = -1.09, R) >droAna3.scaffold_13337 10843533 60 + 23293914 CGACAGAGAUAGCUGUCUGCGGUUGGGCUAACGCUAUCUCAUUCGGUGUGAUUAAGUACG (((..((((((((.((..((......))..))))))))))..)))............... ( -18.30, z-score = -1.58, R) >droYak2.chr3L 4123117 57 - 24197627 AGAUAGAGAUAGCUGUCUAC--GUCCCCUAUCGCUAUCUCACCCCAGGCGAUCAAGUGG- ((((((......))))))..--...((((((((((...........))))))..)).))- ( -11.80, z-score = -0.14, R) >droSec1.super_11 375528 57 + 2888827 AGAUAGAGAUAGCUGUCUAC--CUACCCUAUCGCUAUCUCACCACAGGUGAUUAAGUGG- .(((((.(((((..((....--..)).))))).)))))(((((...)))))........- ( -14.40, z-score = -1.24, R) >droSim1.chr3L 19834366 57 + 22553184 AGAUAGAGAUAGCUGUCUAC--CUACCCUAUCGCUAUCUCACCCCAGGUGAUUAAGUGG- .(((((.(((((..((....--..)).))))).)))))(((((...)))))........- ( -14.40, z-score = -1.36, R) >consensus AGAUAGAGAUAGCUGUCUAC__CUACCCUAUCGCUAUCUCACCCCAGGUGAUUAAGUGG_ .....((((((((...................)))))))).................... ( -6.79 = -7.54 + 0.75)
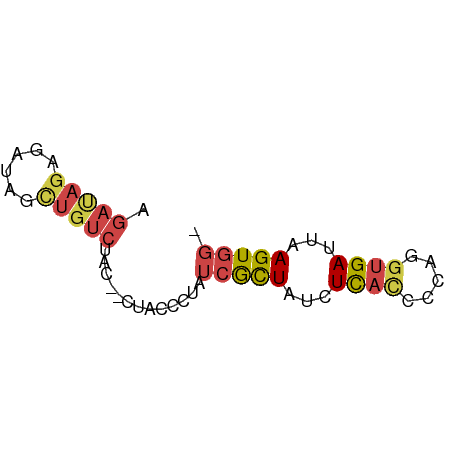
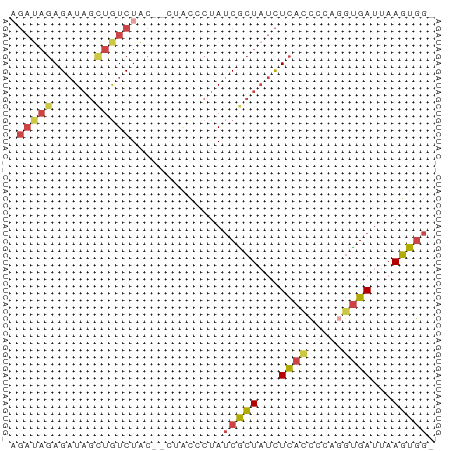
| Location | 20,484,761 – 20,484,818 |
|---|---|
| Length | 57 |
| Sequences | 6 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 70.27 |
| Shannon entropy | 0.58165 |
| G+C content | 0.44664 |
| Mean single sequence MFE | -15.70 |
| Consensus MFE | -6.79 |
| Energy contribution | -8.54 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
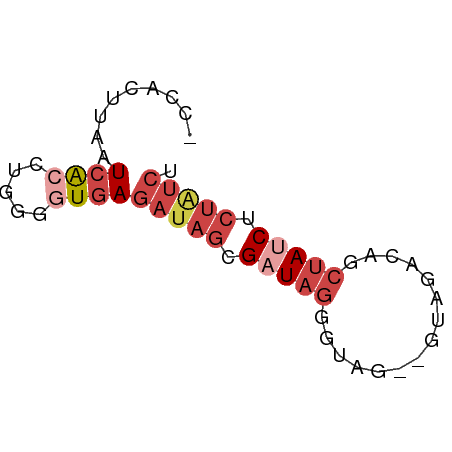
>dm3.chr3L 20484761 57 - 24543557 -CCACUUAAUCACCUGGGGUGAGAUAGCGAUAGGGUCG--GUAGACAACUAUCUCUAUCU -........(((((...)))))(((((.(((((.(((.--...)))..))))).))))). ( -19.60, z-score = -2.55, R) >dp4.chrXR_group8 7892956 52 - 9212921 -CCACUUUAUCAGAAAAGAUAAAGGAAUGAUA-UAUAUAUAUAGAUAUAGAGCU------ -...(((((((......)))))))....(...-(((((......)))))...).------ ( -7.30, z-score = -1.76, R) >droAna3.scaffold_13337 10843533 60 - 23293914 CGUACUUAAUCACACCGAAUGAGAUAGCGUUAGCCCAACCGCAGACAGCUAUCUCUGUCG ...............(((..(((((((((((.((......)).))).))))))))..))) ( -18.00, z-score = -3.90, R) >droYak2.chr3L 4123117 57 + 24197627 -CCACUUGAUCGCCUGGGGUGAGAUAGCGAUAGGGGAC--GUAGACAGCUAUCUCUAUCU -...............((((((((((((..(((....)--.))....)))))))).)))) ( -15.90, z-score = -0.31, R) >droSec1.super_11 375528 57 - 2888827 -CCACUUAAUCACCUGUGGUGAGAUAGCGAUAGGGUAG--GUAGACAGCUAUCUCUAUCU -........(((((...)))))(((((.(((((.((..--....))..))))).))))). ( -16.70, z-score = -1.23, R) >droSim1.chr3L 19834366 57 - 22553184 -CCACUUAAUCACCUGGGGUGAGAUAGCGAUAGGGUAG--GUAGACAGCUAUCUCUAUCU -........(((((...)))))(((((.(((((.((..--....))..))))).))))). ( -16.70, z-score = -1.04, R) >consensus _CCACUUAAUCACCUGGGGUGAGAUAGCGAUAGGGUAG__GUAGACAGCUAUCUCUAUCU .........((((.....))))(((((.(((((...............))))).))))). ( -6.79 = -8.54 + 1.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:59 2011