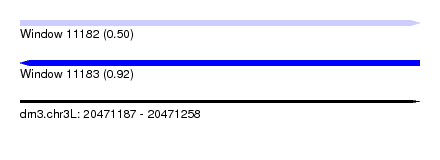
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,471,187 – 20,471,258 |
| Length | 71 |
| Max. P | 0.924008 |

| Location | 20,471,187 – 20,471,258 |
|---|---|
| Length | 71 |
| Sequences | 4 |
| Columns | 87 |
| Reading direction | forward |
| Mean pairwise identity | 50.59 |
| Shannon entropy | 0.77910 |
| G+C content | 0.52708 |
| Mean single sequence MFE | -24.57 |
| Consensus MFE | -5.47 |
| Energy contribution | -8.35 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
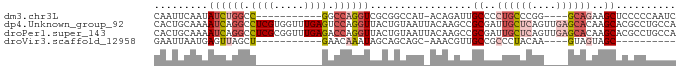
>dm3.chr3L 20471187 71 + 24543557 CAAUUCAAUAUCUGGCC-----------GGCCAGGUCGCGGCCAU-ACAGAUUGCCCCUGCCCGG----GCAGAAGCUCCCCCAAUC ..........((((.((-----------((.((((..(((..(..-...)..))).)))).))))----.))))............. ( -23.80, z-score = -1.08, R) >dp4.Unknown_group_92 4842 87 - 26366 CACUGCAAAAUCAGGCCUCGUGGUUUGAGUCCAGGUUACUGUAAUUACAAGCCGCGAUUGCUCAGUUGAGCACAAGCACGCCUGCCA ...........(((((.(((((((((((((.(((....)))..))).)))))))))).(((((....))))).......)))))... ( -30.90, z-score = -2.40, R) >droPer1.super_143 21433 87 + 108547 CACUGCAAAAUCAGGCCUCGCGGUUUGAGACCAGGUUACUGUAAUUACAAGCCGCGAUUGCUCAGUUGAGCACAAGCACGCCUGCCA ...........(((((.((((((((((.((.(((....)))...)).)))))))))).(((((....))))).......)))))... ( -32.30, z-score = -2.91, R) >droVir3.scaffold_12958 2650419 61 - 3547706 GAAUUAAUGAGUUAGCU-----------GAACAAAUAGCAGCAGC-AAACGUUGCCGCCCUACAA----GUAGUAGC---------- ..............(((-----------(........((.(((((-....))))).)).(((...----.)))))))---------- ( -11.30, z-score = -0.52, R) >consensus CAAUGCAAAAUCAGGCC___________GACCAGGUUACUGCAAU_ACAAGCCGCCACUGCUCAG____GCACAAGCACGCCUGCCA .........((((((.((((.....)))).)))))).................((..((((((....))))))..)).......... ( -5.47 = -8.35 + 2.88)
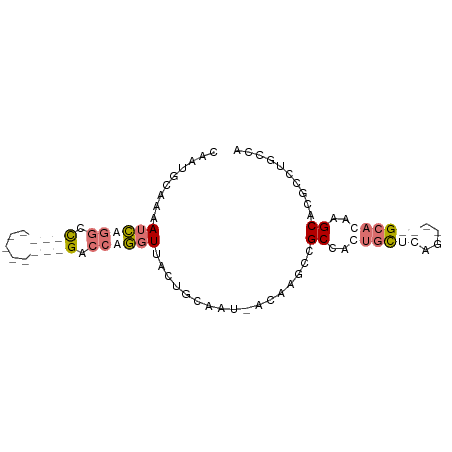
| Location | 20,471,187 – 20,471,258 |
|---|---|
| Length | 71 |
| Sequences | 4 |
| Columns | 87 |
| Reading direction | reverse |
| Mean pairwise identity | 50.59 |
| Shannon entropy | 0.77910 |
| G+C content | 0.52708 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -10.27 |
| Energy contribution | -14.90 |
| Covariance contribution | 4.63 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.924008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20471187 71 - 24543557 GAUUGGGGGAGCUUCUGC----CCGGGCAGGGGCAAUCUGU-AUGGCCGCGACCUGGCC-----------GGCCAGAUAUUGAAUUG ((((.((...((((((((----....)))))))).(((((.-.((((((.....)))))-----------)..))))).)).)))). ( -29.60, z-score = -1.23, R) >dp4.Unknown_group_92 4842 87 + 26366 UGGCAGGCGUGCUUGUGCUCAACUGAGCAAUCGCGGCUUGUAAUUACAGUAACCUGGACUCAAACCACGAGGCCUGAUUUUGCAGUG ..((((((.(((...(((((....)))))...))))))))).......((((...((.(((.......))).)).....)))).... ( -25.00, z-score = -0.01, R) >droPer1.super_143 21433 87 - 108547 UGGCAGGCGUGCUUGUGCUCAACUGAGCAAUCGCGGCUUGUAAUUACAGUAACCUGGUCUCAAACCGCGAGGCCUGAUUUUGCAGUG ...(((((.......(((((....))))).((((((.(((......(((....)))....))).)))))).)))))........... ( -30.30, z-score = -1.27, R) >droVir3.scaffold_12958 2650419 61 + 3547706 ----------GCUACUAC----UUGUAGGGCGGCAACGUUU-GCUGCUGCUAUUUGUUC-----------AGCUAACUCAUUAAUUC ----------(((...((----..(((((((((((.....)-)))))).))))..))..-----------))).............. ( -17.30, z-score = -2.41, R) >consensus UGGCAGGCGUGCUUCUGC____CUGAGCAAUCGCAACUUGU_AUUACAGCAACCUGGUC___________GGCCAGAUUUUGAAGUG ..((((((.(((..((((((....))))))..)))))))))............((((((((.......))))))))........... (-10.27 = -14.90 + 4.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:57 2011