
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,430,232 – 20,430,330 |
| Length | 98 |
| Max. P | 0.999111 |

| Location | 20,430,232 – 20,430,330 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 63.59 |
| Shannon entropy | 0.59014 |
| G+C content | 0.30300 |
| Mean single sequence MFE | -20.26 |
| Consensus MFE | -11.14 |
| Energy contribution | -12.18 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.65 |
| SVM RNA-class probability | 0.999111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
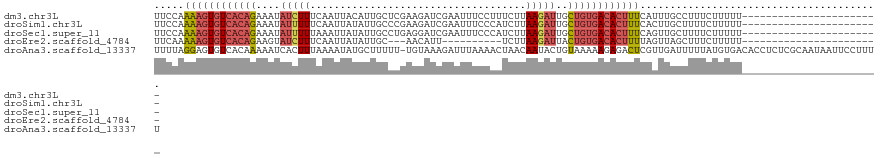
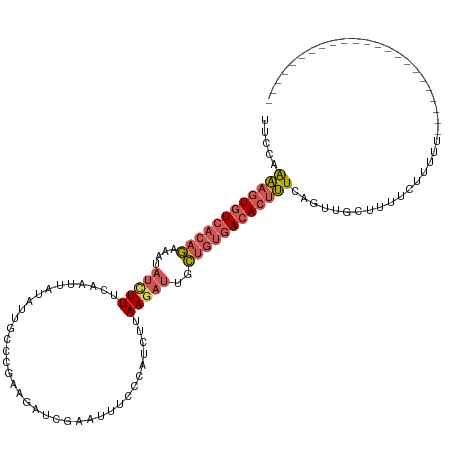
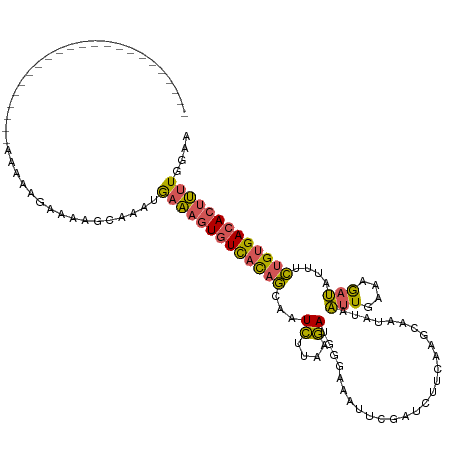
>dm3.chr3L 20430232 98 + 24543557 UUCCAAAAGUGUCACAGAAAUAUCUUUCAAUUACAUUGCUCGAAGAUCGAAUUUCCUUUCUUAAGAUUGCUGUGACACUUUCAUUUGCCUUUCUUUUU----------------------- .....((((((((((((....(((((.............(((.....)))............)))))..)))))))))))).................----------------------- ( -22.41, z-score = -3.55, R) >droSim1.chr3L 19786033 98 + 22553184 UUCCAAAAGUGUCACAGAAAUAUUUUUCAAUUAUAUUGCCCGAAGAUCGAAUUUCCCAUCUUAAGAUUGCUGUGACACUUUCACUUGCUUUUCUUUUU----------------------- .....((((((((((((.(((((.........))))).....(((((.(......).))))).......)))))))))))).................----------------------- ( -19.30, z-score = -2.92, R) >droSec1.super_11 325382 98 + 2888827 UUCCAAAAGUGUCACAGAAAUAUUUUUAAAUUAUAUUGCCUGAGGAUCGAAUUUCCCAUCUUAAGAUUGCUGUGACACUUUCAGUUGCUUUUCUUUUU----------------------- .....((((((((((((.(((((.........)))))..((.(((((.(......).))))).))....)))))))))))).................----------------------- ( -20.20, z-score = -2.13, R) >droEre2.scaffold_4784 20131901 85 + 25762168 UUCAAAAAGUGUCACAGAAGUAUCUUUCAAUUAUAUUGC---AACAUU----------UCUUAAGAUUACUGUGACACUUUUAGUUAGCUUUCUUUUU----------------------- ....(((((((((((((....(((((.............---......----------....)))))..)))))))))))))................----------------------- ( -19.60, z-score = -3.45, R) >droAna3.scaffold_13337 10797660 120 + 23293914 UUUUAGGAGUGUCACAAAAAUCACUUUAAAAUAUGCUUUUU-UGUAAAGAUUUAAAACUAACAAUACUGUAAAAAGAGACUCGUUGAUUUUUAUGUGACACCUCUCGCAAUAAUUCCUUUU ....((..(((((((((((((((.((((((...(((.....-.)))....))))))....(((....)))..............)))))))..))))))))..))................ ( -19.80, z-score = -1.18, R) >consensus UUCCAAAAGUGUCACAGAAAUAUCUUUCAAUUAUAUUGCCCGAAGAUCGAAUUUCCCAUCUUAAGAUUGCUGUGACACUUUCAGUUGCUUUUCUUUUU_______________________ .....((((((((((((....(((((....................................)))))..))))))))))))........................................ (-11.14 = -12.18 + 1.04)
| Location | 20,430,232 – 20,430,330 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 63.59 |
| Shannon entropy | 0.59014 |
| G+C content | 0.30300 |
| Mean single sequence MFE | -22.84 |
| Consensus MFE | -12.10 |
| Energy contribution | -11.50 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.998644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
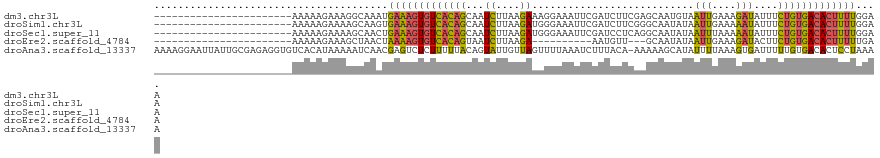
>dm3.chr3L 20430232 98 - 24543557 -----------------------AAAAAGAAAGGCAAAUGAAAGUGUCACAGCAAUCUUAAGAAAGGAAAUUCGAUCUUCGAGCAAUGUAAUUGAAAGAUAUUUCUGUGACACUUUUGGAA -----------------------................(((((((((((((..(((((...........((((.....))))((((...)))).)))))....))))))))))))).... ( -23.10, z-score = -2.17, R) >droSim1.chr3L 19786033 98 - 22553184 -----------------------AAAAAGAAAAGCAAGUGAAAGUGUCACAGCAAUCUUAAGAUGGGAAAUUCGAUCUUCGGGCAAUAUAAUUGAAAAAUAUUUCUGUGACACUUUUGGAA -----------------------................(((((((((((((...(((.(((((.((....)).))))).))).......(((....)))....))))))))))))).... ( -24.60, z-score = -3.06, R) >droSec1.super_11 325382 98 - 2888827 -----------------------AAAAAGAAAAGCAACUGAAAGUGUCACAGCAAUCUUAAGAUGGGAAAUUCGAUCCUCAGGCAAUAUAAUUUAAAAAUAUUUCUGUGACACUUUUGGAA -----------------------..............(..((((((((((((..(((....)))((((.......)))).........................))))))))))))..).. ( -24.40, z-score = -3.23, R) >droEre2.scaffold_4784 20131901 85 - 25762168 -----------------------AAAAAGAAAGCUAACUAAAAGUGUCACAGUAAUCUUAAGA----------AAUGUU---GCAAUAUAAUUGAAAGAUACUUCUGUGACACUUUUUGAA -----------------------..............(.(((((((((((((..(((((..((----------.((((.---...))))..))..)))))....))))))))))))).).. ( -20.10, z-score = -3.24, R) >droAna3.scaffold_13337 10797660 120 - 23293914 AAAAGGAAUUAUUGCGAGAGGUGUCACAUAAAAAUCAACGAGUCUCUUUUUACAGUAUUGUUAGUUUUAAAUCUUUACA-AAAAAGCAUAUUUUAAAGUGAUUUUUGUGACACUCCUAAAA ...((((.......(....)((((((((..(((((((..((((..((((((...(((..(............)..))).-))))))...)))).....))))))))))))))))))).... ( -22.00, z-score = -1.14, R) >consensus _______________________AAAAAGAAAAGCAAAUGAAAGUGUCACAGCAAUCUUAAGAUGGGAAAUUCGAUCUUCAAGCAAUAUAAUUGAAAGAUAUUUCUGUGACACUUUUGGAA .......................................(((((((((((((...((....))...........................(((....)))....))))))))))))).... (-12.10 = -11.50 + -0.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:55 2011