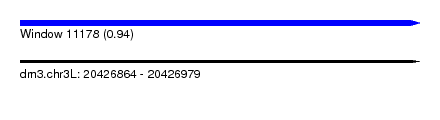
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,426,864 – 20,426,979 |
| Length | 115 |
| Max. P | 0.940642 |

| Location | 20,426,864 – 20,426,979 |
|---|---|
| Length | 115 |
| Sequences | 15 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.65 |
| Shannon entropy | 0.60483 |
| G+C content | 0.44828 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -18.86 |
| Energy contribution | -17.68 |
| Covariance contribution | -1.18 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.940642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
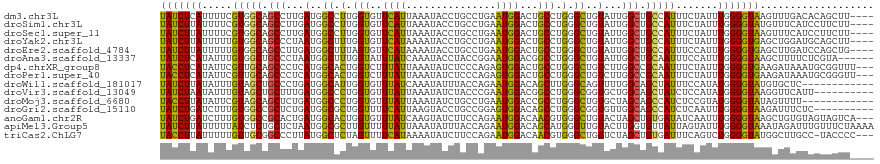
>dm3.chr3L 20426864 115 + 24543557 UAUCUCAUUUUCGUGGCAGCCUUGAUGGCCUUGGUGUUCAUUAAAUACCUGCCUGAAUGGACUGCCUGGGCUGUAUUGGCUGCCAUUUCUAUUUGGGGUAAGUUUGACACAGCUU---- (((((((..(..(((((((((...(((((((.((((((((((.............))))))).))).)))))))...)))))))))....)..)))))))...............---- ( -43.32, z-score = -3.50, R) >droSim1.chr3L 19782282 115 + 22553184 UAUCUUAUUUUCGUGGCAGCCUUGAUGGCCUUGGUGUUCAUUAAAUACCUGCCUGAAUGGACUGCCUGGGCUGUAUUGGCUGCCAUUUCUAUUUGGGGUAUGUUUCAUCCUUCUU---- (((((((..(..(((((((((...(((((((.((((((((((.............))))))).))).)))))))...)))))))))....)..)))))))...............---- ( -41.02, z-score = -3.98, R) >droSec1.super_11 322049 115 + 2888827 UAUCUUAUUUUCGUGGCAGCCUUGAUGGCCUUGGUGUUCAUUAAAUACCUGCCUGAAUGGACUGCCUGGGCUGUAUUGGCUGCCAUUUCUAUUUGGGGUAAGUUUCAUCCUUCUU---- (((((((..(..(((((((((...(((((((.((((((((((.............))))))).))).)))))))...)))))))))....)..)))))))...............---- ( -41.22, z-score = -3.83, R) >droYak2.chr3L 4064765 115 - 24197627 UAUCUUAUUUUUGUGGCAGCCCUAAUGGCUUUGGUGUUCAUAAAAUACCUGCCUGAAUGGACUGCCUGGGCUGUAUUGGCUGCCAUUUCUAUUUGGGGUGAGCUGGAUGCAGCUU---- ..............(((((.((....(((...((((((.....)))))).))).....)).))))).(((((((((..(((..(((..(.....)..))))))..).))))))))---- ( -42.40, z-score = -2.73, R) >droEre2.scaffold_4784 20128564 115 + 25762168 UAUCUUAUUUUUGUGGCAGCCUUGAUGGCUUUAGUGUUCAUAAAAUACCUGCCUGAAUGGACUGCCUGGGCUGUAUUGGCUGCCAUUUCCAUUUGGGGUGAGCUUGAUCCAGCUG---- (((((((.....(((((((((...(((((((..((((((((...............)))))).))..)))))))...))))))))).......)))))))((((......)))).---- ( -37.16, z-score = -1.58, R) >droAna3.scaffold_13337 10794119 113 + 23293914 UAUCUCAUAUUUGUGGCUGCCCUAAUGGCUUUGGUAUUUAUCAAAUACCUACCGGAAUGGACGGCCUGGGCUGUAUUGGCUGCAAUUUCCAUUUGGGGUAAGCUUUUCUCGUA------ ..............(((((((((((.((....((((((.....))))))..))((((..(.(((((...........))))))...))))..))))))).)))).........------ ( -32.20, z-score = -0.79, R) >dp4.chrXR_group8 7846021 116 + 9212921 UACCUCAUAUUCGUUGCAGCCCUCAUGGCACUGGUCUUUAUUAAAUAUCUCCCAGAGUGGACUGCCUGGGCUGUCUUGGCCGCAAUUUCUAUUUGGGGUGAAGAUAAAUGCGGUUU--- .(((.(((........((((((....(((.((((.................))))((....))))).))))))((((.(((.(((.......))).))).))))...))).)))..--- ( -35.13, z-score = -1.03, R) >droPer1.super_40 35776 116 + 810552 UACCUCAUAUUCGUUGCAGCCCUCAUGGCACUGGUCUUUAUUAAAUAUCUCCCAGAGUGGACUGCCUGGGCUGUCUUGGCCGCAAUUUCUAUUUGGGGUGAAGAUAAAUGCGGGUU--- ..((.(((........((((((....(((.((((.................))))((....))))).))))))((((.(((.(((.......))).))).))))...))).))...--- ( -34.93, z-score = -0.60, R) >droWil1.scaffold_181017 529522 107 + 939937 UAUCUUAUAUUUGUAGCUGCCCUGAUGGCAUUGGUGUUUAUCAAAUAUUUACCAGAAUGGACAGCUUGGGCAGUUUUGGCAGCUAUUUCCAUAUGGGGUAUGUGCUC------------ ((((((((((..(((((((((..(((.((...(.((((((((............).))))))).)....)).)))..)))))))))....)))))))))).......------------ ( -32.30, z-score = -1.31, R) >droVir3.scaffold_13049 6324640 109 - 25233164 UAUCUAAUAUUUGUAGCUGCUUUGAUGGCCCUGGUGUUUAUUAAAUAUCUACCCGAAUGGACGGCCUGGGCGGUGCUGGCAGCUAUCUCCAUAUGGGGUAAGGUUCAUU---------- .((((.......(((((((((...((.((((.((((((((((.............))))))).))).)))).))...)))))))))((((....))))..)))).....---------- ( -35.72, z-score = -1.22, R) >droMoj3.scaffold_6680 12316230 107 + 24764193 UACCUUAUAUUCGUAGCAGCUCUGAUGGCCCUGGUGUUUAUUAAAUAUCUGCCUGAAUGGACCGCCUGGGCUGUGCUAGCAGCCAUCUCCGUAUGGGGUAUAGUUUU------------ ((((((((((..((.((.(((...(((((((.((((((((((.............)))))).)))).)))))))...))).)).))....)))))))))).......------------ ( -31.62, z-score = -0.24, R) >droGri2.scaffold_15110 4689470 109 - 24565398 UAUCUGAUCUUUGUGGCCGCUCUGAUGGCGCUGGUUUUCAUUAAGUACCUGCCGGAGUGGACAGCCUGGGCGGUGUUGGCAGCCAUCUCAAUUUGGGGUAAGAUUUCUC---------- .....((((((...(((.(((..((((.((((((((((((((..((....))...)))))).))))..)))).))))))).)))(((((.....)))))))))))....---------- ( -29.60, z-score = 1.34, R) >anoGam1.chr2R 32607706 116 - 62725911 UAUCUGAUCUUUGUGGCCGCACUGAUGGCACUGGUGUUUAUCAAGUAUCUUCCAGAAUGGACAACGUGGGCUGUACUAGCUGUGAUAUCAAUUUGGGGUAAGCUGUGUAGUAGUCA--- .....(((......((((.(((((((((((....)).)))))).......(((.....)))....))))))).((((((((...((..(.....)..)).))))).)))...))).--- ( -23.50, z-score = 2.59, R) >apiMel3.Group5 6707945 119 - 13386189 UAUCUUAUUUUUAUCUCUGCUCUAAUGGCGCUUGUUUUUAUUAAAUAUUUACCAGAAUGGACAGCAUGGGUUGUACUUGGUGUUAUUAGUAUUUGGGGUAAAUAGAUUUGUUUCUAAAA .........((((((((....(((((((((((.((................((.....))(((((....)))))))..))))))))))).....))))))))((((......))))... ( -26.20, z-score = -1.79, R) >triCas2.ChLG7 15742062 115 + 17478683 UACCUUAUUUUUGUUGCGGCCCUUAUGGCUCUAGUUUUCAUAAAAUAUCUUCCAGAAUGGACAACGUGGGCUGUUCUAGCUGUGAUUUCAGUCUGGGGUAUGGCUUGCC-UACCCC--- ...........((..(((((((((((((.........)))))).......(((.....)))......)))))))..))((((......))))..((((((.((....))-))))))--- ( -30.20, z-score = -0.41, R) >consensus UAUCUUAUUUUUGUGGCAGCCCUGAUGGCCCUGGUGUUUAUUAAAUAUCUGCCAGAAUGGACUGCCUGGGCUGUAUUGGCUGCCAUUUCUAUUUGGGGUAAGGUUUAUCCUGCUU____ (((((((.....(((((.(((...(..((.(.(((..((((...............))))...))).).))..)...))).))))).......)))))))................... (-18.86 = -17.68 + -1.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:53 2011