| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,414,082 – 20,414,147 |
| Length | 65 |
| Max. P | 0.602122 |

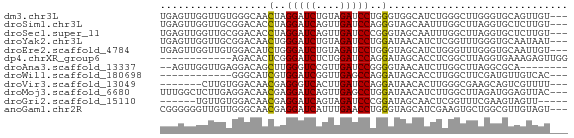
| Location | 20,414,082 – 20,414,147 |
|---|---|
| Length | 65 |
| Sequences | 12 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 63.13 |
| Shannon entropy | 0.80110 |
| G+C content | 0.51124 |
| Mean single sequence MFE | -18.23 |
| Consensus MFE | -5.71 |
| Energy contribution | -5.79 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
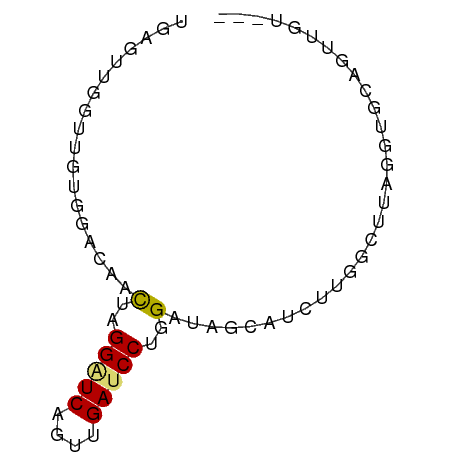
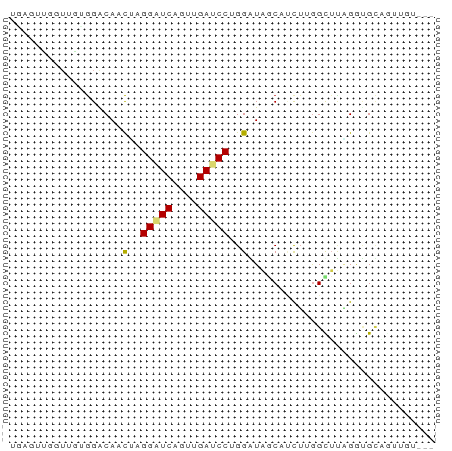
>dm3.chr3L 20414082 65 - 24543557 UGAGUUGGUUGUGGGCAACUAGGAUCUGUAGAUCCUGGGUGGCAUCUGGGCUUGGGUGCAGUUGU--- ..............((((((((((((....)))))).....((((((......))))))))))))--- ( -20.90, z-score = -1.39, R) >droSim1.chr3L 19771505 65 - 22553184 UGAGUUGGUUGCGGACACCUAGGAUCAGUUGAUCCAGGGUAGCAAUUUGGCUUAGGUGCUCUUGU--- (((((..(((((..((.(((.(((((....)))))))))).)))).)..)))))...........--- ( -23.70, z-score = -2.50, R) >droSec1.super_11 311242 65 - 2888827 UGAGUUGGUUGCGGACACCUAGGAUCAGUUGAUCCCGGGUAGCAAUUUGGCUUAGGUGCUCUUGU--- (((((..(((((....(((..(((((....)))))..))).)))).)..)))))...........--- ( -23.60, z-score = -2.19, R) >droYak2.chr3L 4052928 65 + 24197627 UGAGUUGGUUGCGGACAACUGGGAUCUGUAGAUCCUGGAUAACAUCUCGGUUUGGGUGCAAUAAU--- .......((((((..((.(..(((((....)))))..)..(((......)))))..))))))...--- ( -18.40, z-score = -1.57, R) >droEre2.scaffold_4784 20113999 65 - 25762168 UGAGUUGGUUGUGGACAUCUGGGAUCUGUAGAUCCUGGGUAGCAUCUGGGUUUGGGUGCAAUUGU--- ................(((..(((((....)))))..))).((((((......))))))......--- ( -20.60, z-score = -2.13, R) >dp4.chrXR_group6 243848 56 + 13314419 ------------AGACACUCGGGAUCUCUGGAUCCAGGAUAGCACCUCGGCUUAGGUGAAAGAGUUGG ------------...(((((.(((((....))))).......(((((......)))))...))).)). ( -16.80, z-score = -0.56, R) >droAna3.scaffold_13337 10781307 58 - 23293914 --AGUUGGUUGAGGACAGCUUGGGUCCGUUGAUCCGGGGUAACAUCUUGGCUUAGGCGCA-------- --.((((((..(((..(.((((((((....)))))))).)....)))..)))..)))...-------- ( -16.60, z-score = -0.17, R) >droWil1.scaffold_180698 8489440 53 + 11422946 ------------GGGCAUCGUGGAUCGGUUGAGCCAGGAUAGCACCUUGGCUUCGAUGUUGUCAC--- ------------.(((((((..........((((((((.......))))))))))))))).....--- ( -16.70, z-score = -0.86, R) >droVir3.scaffold_13049 13639024 58 - 25233164 -------CUUGUGGACAACGAGGGUCACUUGAUCCAGGAUAACACUUGGGCGAAGCAGUCGUUUU--- -------.........(((((..((...(((.((((((......))))))))).))..)))))..--- ( -12.60, z-score = 0.14, R) >droMoj3.scaffold_6680 11062720 65 + 24764193 UUUGGCUCUUGAGGACAACGAGGAUCAGUUGAGCCUGGAUAACAUCUUGGCUUAGAUGGAGUUAC--- ..((((((((.(((.((((........))))..))).))...(((((......))))))))))).--- ( -17.70, z-score = -1.46, R) >droGri2.scaffold_15110 8974209 57 + 24565398 ------UGUUGUGGACAACGAGGAUCAGUAGAUCCCGGAUAGCAACUCGGUUUCGAAGUAGUU----- ------.(((((......((.(((((....)))))))....)))))(((....))).......----- ( -14.70, z-score = -1.23, R) >anoGam1.chr2R 34780438 65 - 62725911 CGGGGGGUUGUUGGGCAACGAGGAUCAUUUGAACCUGGGUAGCAUCGAAGUGCUGGCGUUGUAGU--- ..............(((((.(((.((....)).))).(.((((((....)))))).))))))...--- ( -16.50, z-score = -0.39, R) >consensus UGAGUUGGUUGUGGACAACUAGGAUCAGUUGAUCCUGGAUAGCAUCUUGGCUUAGGUGCAGUUGU___ ..................(..(((((....)))))..).............................. ( -5.71 = -5.79 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:52 2011