| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,387,436 – 20,387,539 |
| Length | 103 |
| Max. P | 0.872915 |

| Location | 20,387,436 – 20,387,537 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 71.20 |
| Shannon entropy | 0.56347 |
| G+C content | 0.40288 |
| Mean single sequence MFE | -24.31 |
| Consensus MFE | -8.27 |
| Energy contribution | -8.18 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.69 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.699357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
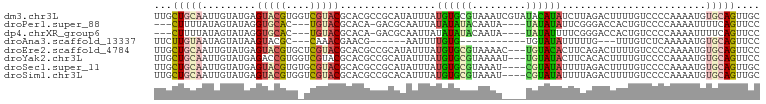
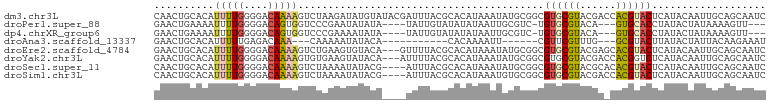
>dm3.chr3L 20387436 101 - 24543557 UUGCUGCAAUUGUAUGAGUACGUGGUCGUACGCACGCCGCAUAUUUAUGUGCGUAAAUCGUAUACAUAUCUUAGACUUUUGUCCCCAAAAUGUGCAGUUGC .....((((((((((..(((..((((..((((((((...........)))))))).))))..)))........(((....)))........)))))))))) ( -28.20, z-score = -1.38, R) >droPer1.super_88 93304 90 + 213009 ---CUUUUAUAGUAUAGGUGCAC---UGUACGCACA-GACGCAAUUAUAUAUACAAUA----UAUAUAUUCGGGACCACUGUCCCCAAAAUUUUCAGUUCC ---........((((((.....)---)))))((...-...))...((((((((...))----))))))...(((((....)))))................ ( -18.30, z-score = -1.93, R) >dp4.chrXR_group6 220760 90 + 13314419 ---CUUUUAUAGUAUAGGUGCAC---UGUACGCACA-GACGCAAUUAUAUAUACAAUA----UAUAUUUUCGGGACCACUGUCCCCAAAAUUUUCAGUUCC ---........((((((.....)---)))))((...-...))...(((((((...)))----))))((((.(((((....))))).))))........... ( -17.90, z-score = -1.96, R) >droAna3.scaffold_13337 10754582 78 - 23293914 UUCUUGUAAUAGUAUAAGUACGC---CAAACGAACG------AAUUUUGUG-----------UGUAUAUUUUUG---UUUGUCUCAAAAAUGUGCAGUUCC ..((((((....)))))).....---...(((((..------...))))).-----------((((((((((((---.......))))))))))))..... ( -13.90, z-score = -1.52, R) >droEre2.scaffold_4784 20087429 98 - 25762168 UUGCUGCAAUUGUAUGAGUACGUGCUCGUACGCACGCCGCAUAUUUAUGUGCGUAAAAC---UGUACACUUCAGACUUUUGUCCCCAAAAUGUGCAGUUCC ..(((((....((((((((....))))))))))).))((((((....))))))...(((---((((((.....(((....))).......))))))))).. ( -30.10, z-score = -2.97, R) >droYak2.chr3L 4024712 98 + 24197627 UUGCUGCAAUUGUAUGAGACCGUGGUCGUACGCACGCCGCAUAUUUAUGUGCGUAAAAU---UGUAUACUUCACACUUUUGUCCCCAAAAUGUGCAGUUCC ..(((((....(((((.(((....))).((((((((...........))))))))....---..)))))...(((.(((((....)))))))))))))... ( -24.80, z-score = -1.29, R) >droSec1.super_11 281411 97 - 2888827 UUGCUGCAAUUGUAUGAGUACGUGUGCGUACGCACGCCGCAUAUUUAUGUGCGUAAAU----CGUAUAUUUUAGACUUUUGUCCCCAAAAUGUGCAGUUGC .....((((((((((..((((((((((....))))))((((((....)))))).....----.))))......(((....)))........)))))))))) ( -31.00, z-score = -2.57, R) >droSim1.chr3L 19743956 97 - 22553184 UUGCUGCAAUUGUAUGAGUACGUGGUCGUACGCACGCCGCACAUUUAUGUGCGUAAAU----CGUAUAUUUUAGACUUUUGUCCCCAAAAUGUGCAGUUGC .....(((((((..((.(((((....))))).))..).(((((((((((((((.....----)))))))....(((....)))....)))))))))))))) ( -30.30, z-score = -2.27, R) >consensus UUGCUGCAAUUGUAUGAGUACGUG_UCGUACGCACGCCGCAUAUUUAUGUGCGUAAAA____UGUAUAUUUUAGACUUUUGUCCCCAAAAUGUGCAGUUCC ...(((((.........(((((....)))))................((((((.........))))))........................))))).... ( -8.27 = -8.18 + -0.09)
| Location | 20,387,437 – 20,387,539 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 70.27 |
| Shannon entropy | 0.58341 |
| G+C content | 0.39430 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -8.18 |
| Energy contribution | -8.16 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
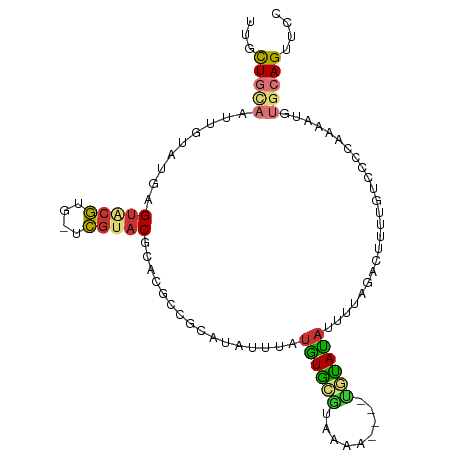
>dm3.chr3L 20387437 102 + 24543557 CAACUGCACAUUUUGGGGACAAAAGUCUAAGAUAUGUAUACGAUUUACGCACAUAAAUAUGCGGCGUGCGUACGACCACGUACUCAUACAAUUGCAGCAAUC ...(((((..(((((....)))))..........(((((........((((........))))(.((((((......)))))).))))))..)))))..... ( -25.70, z-score = -1.90, R) >droPer1.super_88 93305 91 - 213009 GAACUGAAAAUUUUGGGGACAGUGGUCCCGAAUAUAUA----UAUUGUAUAUAUAAUUGCGUC-UGUGCGUACA---GUGCACCUAUACUAUAAAAGUU--- .((((.......((.(((((....))))).))......----.....((((((((..((((.(-(((....)))---)))))..)))).))))..))))--- ( -19.70, z-score = -0.85, R) >dp4.chrXR_group6 220761 91 - 13314419 GAACUGAAAAUUUUGGGGACAGUGGUCCCGAAAAUAUA----UAUUGUAUAUAUAAUUGCGUC-UGUGCGUACA---GUGCACCUAUACUAUAAAAGUU--- .((((....(((((.(((((....))))).)))))...----.....((((((((..((((.(-(((....)))---)))))..)))).))))..))))--- ( -22.80, z-score = -2.21, R) >droAna3.scaffold_13337 10754583 79 + 23293914 GAACUGCACAUUUUUGAGACAAA---CAAAAAUAUACA-----------CACAAAAUU------CGUUCGUUUG---GCGUACUUAUACUAUUACAAGAAAU ..........((((((...((((---(...........-----------.........------.....)))))---..(((....))).....)))))).. ( -4.81, z-score = 1.07, R) >droEre2.scaffold_4784 20087430 99 + 25762168 GAACUGCACAUUUUGGGGACAAAAGUCUGAAGUGUACA---GUUUUACGCACAUAAAUAUGCGGCGUGCGUACGAGCACGUACUCAUACAAUUGCAGCAAUC ((((((.(((((((..((((....))))))))))).))---))))...(((......((((..((((((......))))))...))))....)))....... ( -29.00, z-score = -2.00, R) >droYak2.chr3L 4024713 99 - 24197627 GAACUGCACAUUUUGGGGACAAAAGUGUGAAGUAUACA---AUUUUACGCACAUAAAUAUGCGGCGUGCGUACGACCACGGUCUCAUACAAUUGCAGCAAUC ...(((((..(((((....)))))((((((........---....(((((((.............))))))).(((....)))))))))...)))))..... ( -30.12, z-score = -3.04, R) >droSec1.super_11 281412 98 + 2888827 CAACUGCACAUUUUGGGGACAAAAGUCUAAAAUAUACG----AUUUACGCACAUAAAUAUGCGGCGUGCGUACGCACACGUACUCAUACAAUUGCAGCAAUC ...(((((.....(((((((....))).......((((----.....((((........))))..((((....)))).))))))))......)))))..... ( -24.90, z-score = -2.57, R) >droSim1.chr3L 19743957 98 + 22553184 CAACUGCACAUUUUGGGGACAAAAGUCUAAAAUAUACG----AUUUACGCACAUAAAUGUGCGGCGUGCGUACGACCACGUACUCAUACAAUUGCAGCAAUC ...(((((..(((((....)))))..............----.....((((((....))))))(..((.(((((....))))).))..)...)))))..... ( -27.60, z-score = -2.87, R) >consensus GAACUGCACAUUUUGGGGACAAAAGUCUAAAAUAUACA____UUUUACGCACAUAAAUAUGCGGCGUGCGUACGA_CACGUACUCAUACAAUUGCAGCAAUC ..........(((((....))))).........................................((((((......))))))................... ( -8.18 = -8.16 + -0.01)

| Location | 20,387,437 – 20,387,539 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 70.27 |
| Shannon entropy | 0.58341 |
| G+C content | 0.39430 |
| Mean single sequence MFE | -23.99 |
| Consensus MFE | -9.52 |
| Energy contribution | -9.40 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
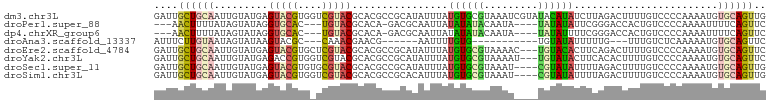
>dm3.chr3L 20387437 102 - 24543557 GAUUGCUGCAAUUGUAUGAGUACGUGGUCGUACGCACGCCGCAUAUUUAUGUGCGUAAAUCGUAUACAUAUCUUAGACUUUUGUCCCCAAAAUGUGCAGUUG ....((((((..((((((((((((....)))))..((((.((((....))))))))...))))))).........(((....))).........)))))).. ( -27.60, z-score = -1.33, R) >droPer1.super_88 93305 91 + 213009 ---AACUUUUAUAGUAUAGGUGCAC---UGUACGCACA-GACGCAAUUAUAUAUACAAUA----UAUAUAUUCGGGACCACUGUCCCCAAAAUUUUCAGUUC ---..........((((((.....)---)))))((...-...))...((((((((...))----))))))...(((((....)))))............... ( -18.30, z-score = -2.03, R) >dp4.chrXR_group6 220761 91 + 13314419 ---AACUUUUAUAGUAUAGGUGCAC---UGUACGCACA-GACGCAAUUAUAUAUACAAUA----UAUAUUUUCGGGACCACUGUCCCCAAAAUUUUCAGUUC ---..........((((((.....)---)))))((...-...))...(((((((...)))----))))((((.(((((....))))).)))).......... ( -17.90, z-score = -2.03, R) >droAna3.scaffold_13337 10754583 79 - 23293914 AUUUCUUGUAAUAGUAUAAGUACGC---CAAACGAACG------AAUUUUGUG-----------UGUAUAUUUUUG---UUUGUCUCAAAAAUGUGCAGUUC ....((((((....)))))).....---...(((((..------...))))).-----------((((((((((((---.......)))))))))))).... ( -13.90, z-score = -1.26, R) >droEre2.scaffold_4784 20087430 99 - 25762168 GAUUGCUGCAAUUGUAUGAGUACGUGCUCGUACGCACGCCGCAUAUUUAUGUGCGUAAAAC---UGUACACUUCAGACUUUUGUCCCCAAAAUGUGCAGUUC ....(((((....((((((((....))))))))))).))((((((....))))))...(((---((((((.....(((....))).......))))))))). ( -30.10, z-score = -2.55, R) >droYak2.chr3L 4024713 99 + 24197627 GAUUGCUGCAAUUGUAUGAGACCGUGGUCGUACGCACGCCGCAUAUUUAUGUGCGUAAAAU---UGUAUACUUCACACUUUUGUCCCCAAAAUGUGCAGUUC ....(((((....(((((.(((....))).((((((((...........))))))))....---..)))))...(((.(((((....))))))))))))).. ( -24.80, z-score = -0.97, R) >droSec1.super_11 281412 98 - 2888827 GAUUGCUGCAAUUGUAUGAGUACGUGUGCGUACGCACGCCGCAUAUUUAUGUGCGUAAAU----CGUAUAUUUUAGACUUUUGUCCCCAAAAUGUGCAGUUG ....((((((..(((((((.(((((((((....)))))).(((((....))))))))..)----)))))).....(((....))).........)))))).. ( -30.50, z-score = -2.62, R) >droSim1.chr3L 19743957 98 - 22553184 GAUUGCUGCAAUUGUAUGAGUACGUGGUCGUACGCACGCCGCACAUUUAUGUGCGUAAAU----CGUAUAUUUUAGACUUUUGUCCCCAAAAUGUGCAGUUG ....((((((..((((((((((((....)))))......((((((....))))))....)----)))))).....(((....))).........)))))).. ( -28.80, z-score = -1.94, R) >consensus GAUUGCUGCAAUUGUAUGAGUACGUG_UCGUACGCACGCCGCAUAUUUAUGUGCGUAAAA____UGUAUAUUUUAGACUUUUGUCCCCAAAAUGUGCAGUUC ....((((((.........(((((....)))))................((((((.........))))))........................)))))).. ( -9.52 = -9.40 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:48 2011