| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,382,537 – 20,382,628 |
| Length | 91 |
| Max. P | 0.834168 |

| Location | 20,382,537 – 20,382,628 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 58.77 |
| Shannon entropy | 0.70363 |
| G+C content | 0.35380 |
| Mean single sequence MFE | -17.42 |
| Consensus MFE | -6.44 |
| Energy contribution | -5.84 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
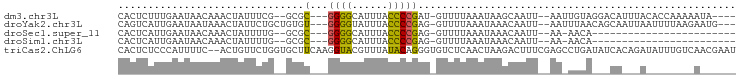
>dm3.chr3L 20382537 91 + 24543557 ----UAUUUUUGGUGUAAAUGUCCUACAAUU--AAUUGCUUAUUUAAAAC-AUCGGGGUAAAUGCCCC---GCGC--CGAAAUAGUUUGUUAUUCAAAGAGUG ----...(((((((((.............((--((........))))...-...(((((....)))))---))))--)))))...((((.....))))..... ( -19.20, z-score = -1.25, R) >droYak2.chr3L 4019759 94 - 24197627 ---CAUUCUUAAAAUUAAUUGCUGUUAAAUU--AAUUGUUUAUUUAAAAC-CUCGGGGUAAAUACCCC---ACACAGCAGAAUAGUUUAUUAUUCAAUGACUG ---((((............(((((((((((.--....)))))........-...(((((....)))))---..))))))((((((....)))))))))).... ( -20.10, z-score = -3.09, R) >droSec1.super_11 276385 70 + 2888827 ------------------------UGUU-UU--AAUUGUUUAUUUAAAAC-CUCGGGGUAAAUGCCCC---GCGC--CAAAAUAGUUUGUUAUUCAAUGAGUG ------------------------.(((-((--((........)))))))-((((((((....)))))---....--((((....)))).........))).. ( -14.70, z-score = -1.62, R) >droSim1.chr3L 19738940 70 + 22553184 ------------------------UGUU-UU--AAUUGUUUAUUUAAAAC-CUCGGGGUAAAUGCCCC---GCGC--CAAAAUAGUUUGUUAUUCAAUGAGUG ------------------------.(((-((--((........)))))))-((((((((....)))))---....--((((....)))).........))).. ( -14.70, z-score = -1.62, R) >triCas2.ChLG6 4396166 101 - 13544221 AUUCGUUGACAAAUAUCUGUGAUAUCAGGCUCGAAAGUCUUAGUUGAGACACCCUGUAUAAACGUACCUUGAAGCACCAGAACAGU--GAAAAUGGGAGAGUG .((((((..((....((((((...(((((.......(((((....))))).....((((....)))))))))..)).))))....)--)..))))))...... ( -18.40, z-score = 0.66, R) >consensus ______U________U___UG___UGAA_UU__AAUUGUUUAUUUAAAAC_CUCGGGGUAAAUGCCCC___GCGC__CAAAAUAGUUUGUUAUUCAAUGAGUG ......................................................(((((....)))))................................... ( -6.44 = -5.84 + -0.60)



| Location | 20,382,537 – 20,382,628 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 58.77 |
| Shannon entropy | 0.70363 |
| G+C content | 0.35380 |
| Mean single sequence MFE | -16.62 |
| Consensus MFE | -5.98 |
| Energy contribution | -5.54 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20382537 91 - 24543557 CACUCUUUGAAUAACAAACUAUUUCG--GCGC---GGGGCAUUUACCCCGAU-GUUUUAAAUAAGCAAUU--AAUUGUAGGACAUUUACACCAAAAAUA---- .........................(--(.((---((((......)))))((-((((((..(((....))--)....))))))))...).)).......---- ( -16.30, z-score = -1.16, R) >droYak2.chr3L 4019759 94 + 24197627 CAGUCAUUGAAUAAUAAACUAUUCUGCUGUGU---GGGGUAUUUACCCCGAG-GUUUUAAAUAAACAAUU--AAUUUAACAGCAAUUAAUUUUAAGAAUG--- ...((...(((((......)))))((((((.(---(((((....))))))..-((((.....))))....--......))))))...........))...--- ( -20.20, z-score = -2.29, R) >droSec1.super_11 276385 70 - 2888827 CACUCAUUGAAUAACAAACUAUUUUG--GCGC---GGGGCAUUUACCCCGAG-GUUUUAAAUAAACAAUU--AA-AACA------------------------ ..(((.........((((....))))--....---((((......)))))))-(((((((........))--))-))).------------------------ ( -15.10, z-score = -2.53, R) >droSim1.chr3L 19738940 70 - 22553184 CACUCAUUGAAUAACAAACUAUUUUG--GCGC---GGGGCAUUUACCCCGAG-GUUUUAAAUAAACAAUU--AA-AACA------------------------ ..(((.........((((....))))--....---((((......)))))))-(((((((........))--))-))).------------------------ ( -15.10, z-score = -2.53, R) >triCas2.ChLG6 4396166 101 + 13544221 CACUCUCCCAUUUUC--ACUGUUCUGGUGCUUCAAGGUACGUUUAUACAGGGUGUCUCAACUAAGACUUUCGAGCCUGAUAUCACAGAUAUUUGUCAACGAAU ...............--.....(((((((((....))))).......((((.(((((......)))).....).))))......)))).(((((....))))) ( -16.40, z-score = 1.06, R) >consensus CACUCAUUGAAUAACAAACUAUUUUG__GCGC___GGGGCAUUUACCCCGAG_GUUUUAAAUAAACAAUU__AA_AACA___CA___A________A______ ...................................((((......))))...................................................... ( -5.98 = -5.54 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:46 2011