| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,303,628 – 20,303,751 |
| Length | 123 |
| Max. P | 0.924341 |

| Location | 20,303,628 – 20,303,751 |
|---|---|
| Length | 123 |
| Sequences | 7 |
| Columns | 129 |
| Reading direction | forward |
| Mean pairwise identity | 61.69 |
| Shannon entropy | 0.68699 |
| G+C content | 0.47326 |
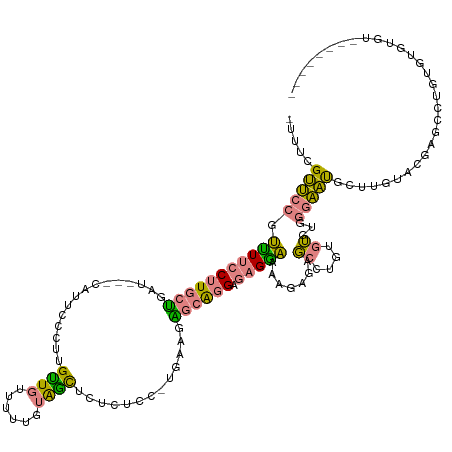
| Mean single sequence MFE | -37.37 |
| Consensus MFE | -7.74 |
| Energy contribution | -6.60 |
| Covariance contribution | -1.13 |
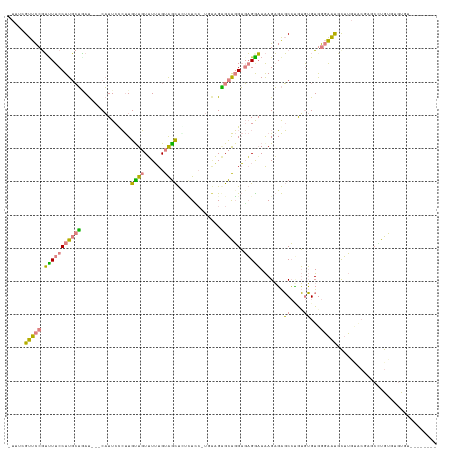
| Combinations/Pair | 1.64 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
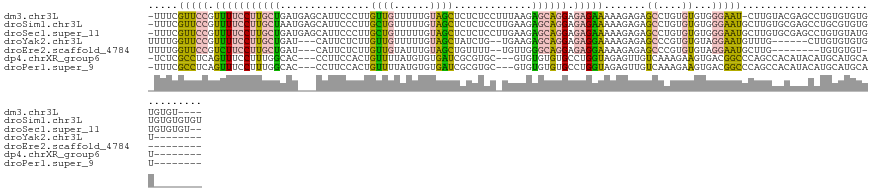
>dm3.chr3L 20303628 123 + 24543557 -UUUCGUUCCGUUUUCCUUGCUGAUGAGCAUUCCCUUGUUGUUUUUGUAGCUCUCUCCUUUAAGAGCAGGAGAGAAAAAGAGAGCCUGUGUGUGGGAAU-CUUGUACGAGCCUGUGUGUGUGUGU---- -....((((.((((((...((((..(((((.........)))))...))))((((((((........))))))))....))))))....(((..((...-))..)))))))..............---- ( -30.50, z-score = -0.89, R) >droSim1.chr3L 19674636 128 + 22553184 -UUUCGUUCCGUUUUCCUUGCUAAUGAGCAUUCCCUUGCUGUUUUUGUAGCUCUCUCCUUGAAGAGCAGGAGAGAAAAAGAGAGCCUGUGUGUGGGAAUGCUUGUGCGAGCCUGCGUGUGUGUGUGUGU -...((..((((....(((((..(..(((((((((.(((.((((((.....((((((((........))))))))....))))))..)))...)))))))))..))))))...))).)..))....... ( -42.60, z-score = -3.47, R) >droSec1.super_11 211937 126 + 2888827 -UUUCGUUCCGUUUUCCUUGCUGAUGAGCAUUCCCUUGCUGUUUUUGUAGCUCUCUCCUUGAAGAGCAGGAGAGAAAAAGAGAGCCUGUGUGUGGGAAUGCUUGUGCGAGCCUGUGUAUGUGUGUGU-- -...............(((((..(..(((((((((.(((.((((((.....((((((((........))))))))....))))))..)))...)))))))))..)))))).................-- ( -40.00, z-score = -3.08, R) >droYak2.chr3L 3941406 110 - 24197627 UUUUGGUUCCGUUUUCCUUGCUGAU---CAUUCUCUUGUUGUUUUUGUAGCUAUCUG--UGAAGAGCAGGAGAGGAAAAGAGAGCCCGUGUGUAGGAAUGUUUG------CUUGUGUGUGU-------- ....(((((..((((((.((.....---)).((((((((..((((..(((....)))--..))))))))))))))))))..)))))((..(..(((........------)))..)..)).-------- ( -28.20, z-score = -1.58, R) >droEre2.scaffold_4784 20027576 106 + 25762168 UUUUGGUUCCGUCUUCCUUGCUGAU---CAUUCUCUUGUUGUAUUUGUAGCUGUUUU--UGUUGGGCAGGAGAGGAAAAGAGAGCCCGUGUGUAGGAAUGCUUG--------UGUGUGU---------- ....((((((.(((((((((((.(.---((.......((((......))))......--)).).)))))).)))))...).)))))((..(..(((....))).--------.)..)).---------- ( -26.32, z-score = -0.88, R) >dp4.chrXR_group6 2161010 114 + 13314419 -UCUCGCCUCAGUUUCCUUUGGCAC---CCUUCCACUGUUUUAUGUGUGAUCGCGUGC---GUGUGUGUGCCUGGUAGAGUUGUCAAAGAAGUGACGGCCCAGCCACAUACAUGCAUGCAU-------- -....(((..((....))..)))..---...(((((........))).))..((((((---(((((((((.((((....(((((((......))))))))))).)))))))))))))))..-------- ( -47.00, z-score = -4.64, R) >droPer1.super_9 454087 114 + 3637205 -UUUCGCCUCAGUUUCCUUUGGCAC---CCUUCCACUGUUUUAUGUGUGAUCGCGUGC---GUGUGUGUGCCUGGUAGAGUUGUCAAAGAAGUGACGGCCCAGCCACAUACAUGCAUGCAU-------- -....(((..((....))..)))..---...(((((........))).))..((((((---(((((((((.((((....(((((((......))))))))))).)))))))))))))))..-------- ( -47.00, z-score = -4.77, R) >consensus _UUUCGUUCCGUUUUCCUUGCUGAU___CAUUCCCUUGUUGUUUUUGUAGCUCUCUCC_UGAAGAGCAGGAGAGGAAAAGAGAGCCUGUGUGUGGGAAUGCUUGUACGAGCCUGUGUGUGU________ .....(((((.(((((((((((...............((((......)))).............)))))).))))).......((....))...))))).............................. ( -7.74 = -6.60 + -1.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:39 2011