| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,244,580 – 20,244,671 |
| Length | 91 |
| Max. P | 0.954607 |

| Location | 20,244,580 – 20,244,671 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 73.78 |
| Shannon entropy | 0.48513 |
| G+C content | 0.44224 |
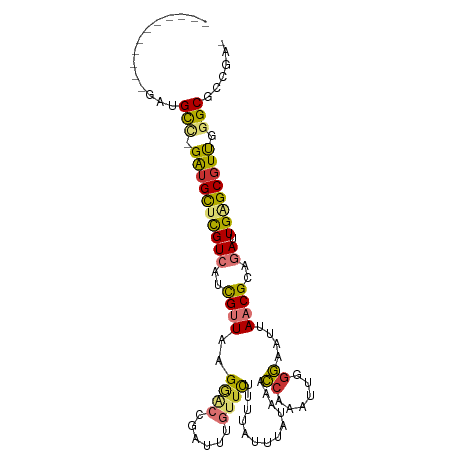
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.18 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
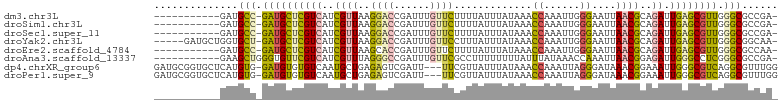
>dm3.chr3L 20244580 91 - 24543557 -----------GAUGCC-GAUGCUCGUCAUCGUUAAGGACCGAUUUGUUCUUUUAUUUAUAAACCAAAUUGGGAAUUAACGCAGAUUGAGCGUUGGGCGCCGA- -----------(.((((-((((((((((..((((((...((((((((.................))))))))...))))))..)).)))))))).)))).)..- ( -30.03, z-score = -2.77, R) >droSim1.chr3L 19615693 91 - 22553184 -----------GAUGCC-GAUGCUCGUCAUCGUUAAGGACCGAUUUGUUCUUUUAUUUAUAAACCAAAUUGGGAAUUAACGCAGAUUGAGCGUUGGGCGCCGA- -----------(.((((-((((((((((..((((((...((((((((.................))))))))...))))))..)).)))))))).)))).)..- ( -30.03, z-score = -2.77, R) >droSec1.super_11 149222 91 - 2888827 -----------GAUGCC-GAUGCUCGUCAUCGUUAAGGACCGAUUUGUUCUUUUAUUUAUAAACCAAAUUGGGAAUUAACGCAGAUUGAGCGUUGGGCGCCGA- -----------(.((((-((((((((((..((((((...((((((((.................))))))))...))))))..)).)))))))).)))).)..- ( -30.03, z-score = -2.77, R) >droYak2.chr3L 3869854 97 + 24197627 -----GAUGCUGGUGCU-GAUGCUCGUCAUCGUUAAGGACCGAUUUGUUCCUUUAUUUAUAAACCAAAUUGGGAAUUAACGCAGAUUGAGCGUUGGGCGGCAA- -----..(((((...(.-.(((((((((..((((((...((((((((.................))))))))...))))))..)).)))))))..).))))).- ( -32.83, z-score = -2.90, R) >droEre2.scaffold_4784 19969606 91 - 25762168 -----------GAUGCC-GAUGCUCGUCAUCGUUAAGCACCGAUUUGUUCUUUUAUUUAUAAACCAAAUUGGGAAUUAACGCAGAUUGAGCGUUGGGCGCCAA- -----------(.((((-((((((((((..((((((...((((((((.................))))))))...))))))..)).)))))))).)))).)..- ( -30.03, z-score = -3.14, R) >droAna3.scaffold_13337 6368936 92 - 23293914 -----------GAAGCUGGGUGUUCGUCAUCGUUUAGGGCCGAUUUGUUCGCCUUUUUUUUAUUUAUAAACCAAAUUAACGGAGAUUGGGCCUCGGGCGCCGA- -----------.......((((((((.........(((.(((((((............((((....))))((........))))))))).)))))))))))..- ( -21.80, z-score = 0.86, R) >dp4.chrXR_group6 2109826 100 - 13314419 GAUGCGGUGCUCAUGUG-GAUGUGUGUCAAUGCUGAGAGUCGAUU---UUCGUUAUUUAUAAACCAAAUUAGGGAUAAACGGAAAUUGGGCGUCAGGCGUUUGG (((((((..(.(((...-.))).)..))....((((..(((((((---(((((((((..(((......)))..))).)))))))))).))).)))))))))... ( -26.90, z-score = -1.34, R) >droPer1.super_9 402990 100 - 3637205 GAUGCGGUGCUCAUGUG-GAUGUGUGUCAAUGCUGAGAGUCGAUU---UUCGUUAUUUAUAAACCAAAUUAGGGAUAAACGGAAAUUGGGCGUCAGGCGUUUGG (((((((..(.(((...-.))).)..))....((((..(((((((---(((((((((..(((......)))..))).)))))))))).))).)))))))))... ( -26.90, z-score = -1.34, R) >consensus ___________GAUGCC_GAUGCUCGUCAUCGUUAAGGACCGAUUUGUUCUUUUAUUUAUAAACCAAAUUGGGAAUUAACGCAGAUUGAGCGUUGGGCGCCGA_ ..............(((.((((((((((..((((..((((......)))).............((......))....))))..)).)))))))).)))...... (-15.28 = -15.18 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:31 2011