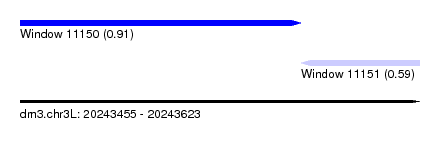
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,243,455 – 20,243,623 |
| Length | 168 |
| Max. P | 0.911770 |

| Location | 20,243,455 – 20,243,573 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.27 |
| Shannon entropy | 0.13507 |
| G+C content | 0.45425 |
| Mean single sequence MFE | -35.26 |
| Consensus MFE | -27.76 |
| Energy contribution | -27.60 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
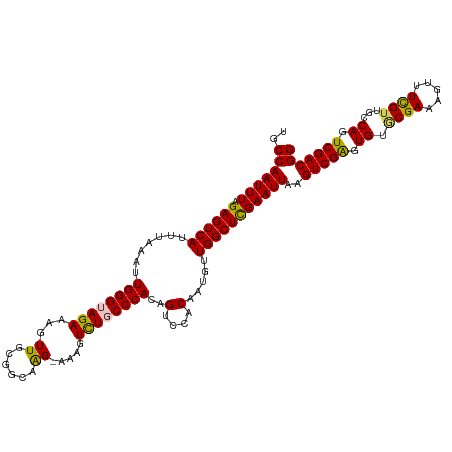
>dm3.chr3L 20243455 118 + 24543557 UGCCAAUUUAGAGUCAUUUAAAUUGGGUAGAAAGUUGCGGCAAGAAAAGUCUGCCCACAGUCCACAAUGUUGGCUCGAAUUAAGUCGGGUGUUCGAAAGUUUUGUUGCCAGUCGACGG ..((((((((((....)))))))))).......((((((((((.((((....(((.(((........))).)))((((((..........))))))...)))).))))).).)))).. ( -33.90, z-score = -1.84, R) >droEre2.scaffold_4784 19968491 115 + 25762168 UGCCAAUUUAGAGUCAUUUAAAUUGGGUAGAAAGUUGCGGUAAG---AUUUUCCCCACAGUCAACAAUGUUGGCUCGAAUUAAGUCGAGUGUACGAAAGUUUCGUUGCCAGUCGACGG ..((((((((((....)))))))))).......(((((((((((---((((((..(((.((((((...))))))((((......)))))))...)))))))...))))).).)))).. ( -32.10, z-score = -2.51, R) >droYak2.chr3L 3868620 112 - 24197627 UGCCAAUUUAGAGUCAUUUAAAUUGGGUAGAAAGUUGCGGUAGG---AUUCUCCCCAAAGUCCACAA---UGGCUCGAAUUAAGUCGAGUGUGCGAAAGUUUCGUUGCCAGUCGACGG ..(((((((.((((((((....(((((.((((............---.)))).))))).......))---)))))))))))..(((((.((.((((........)))))).))))))) ( -31.82, z-score = -2.05, R) >droSec1.super_11 148167 118 + 2888827 UGCCAAUUUCGAGUCAUUUAAAUUGGGUAGAAAGUUGCGGCAAGGAAAGUCUGCCCACAGUCCACAAUGUUGGCUUGAAUUAAGUCGAGUGUGCGAAAGUUUCGUUGCCAGUCGACGG ..((...(((((((((........(((((((...((.(.....).))..)))))))(((........))))))))))))....(((((.((.((((........)))))).))))))) ( -38.20, z-score = -2.69, R) >droSim1.chr3L 19614614 118 + 22553184 UGCCAAUUUCGAGUCAUUUAAAUUGGGUAGAAAGUUGCGGCAAGGAAAGUCUGCCCACAGUCCACAAUGUUGGCUCGAAUUAAGUCGAGUGUGCGAAAGUUUCGUUGCCAGUCGACGG ..((...(((((((((........(((((((...((.(.....).))..)))))))(((........))))))))))))....(((((.((.((((........)))))).))))))) ( -40.30, z-score = -3.30, R) >consensus UGCCAAUUUAGAGUCAUUUAAAUUGGGUAGAAAGUUGCGGCAAG_AAAGUCUGCCCACAGUCCACAAUGUUGGCUCGAAUUAAGUCGAGUGUGCGAAAGUUUCGUUGCCAGUCGACGG ..(((((((.((((((.......((((((((......(.....).....))))))))..(....).....)))))))))))..(((((.((.((((.....))))...)).))))))) (-27.76 = -27.60 + -0.16)

| Location | 20,243,573 – 20,243,623 |
|---|---|
| Length | 50 |
| Sequences | 6 |
| Columns | 50 |
| Reading direction | reverse |
| Mean pairwise identity | 78.13 |
| Shannon entropy | 0.43051 |
| G+C content | 0.32333 |
| Mean single sequence MFE | -9.23 |
| Consensus MFE | -4.57 |
| Energy contribution | -4.47 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589830 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
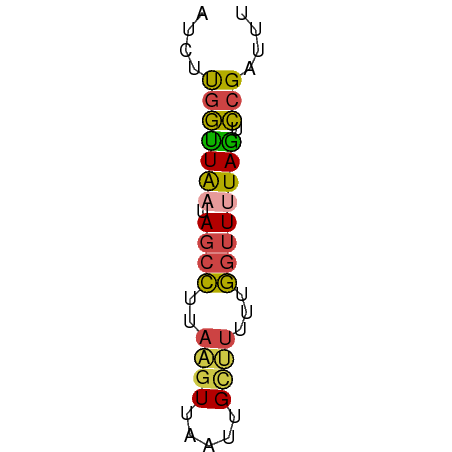
>dm3.chr3L 20243573 50 - 24543557 AUCUUGGUUAUCAGCCUUAAGUUAAUUGCUUUUCGGUUUUAGUCCGAUUU ...((((.....((((..((((.....))))...)))).....))))... ( -8.50, z-score = -1.71, R) >droSim1.chr3L 19614732 50 - 22553184 AUCUUGGUUAAUAGCCUUAAGUUAAUUGCUUUUUGGUUUUAGUCCGAUUU ...((((((((.((((..((((.....))))...)))))))).))))... ( -10.30, z-score = -2.39, R) >droSec1.super_11 148285 50 - 2888827 AUCCUGGUUAAUAGCCUUAAGUUAAUUGCUUUUUGGUUUUAGUCCGAUUU ....(((((((.((((..((((.....))))...)))))))).))).... ( -9.20, z-score = -1.70, R) >droYak2.chr3L 3868732 50 + 24197627 AUCUUGGUUAAUAGCCUUAAAUUAAUUGCUUUUUGGUUUUAGUCCGAUUU ...((((((((.((((..(((........)))..)))))))).))))... ( -7.40, z-score = -1.07, R) >droEre2.scaffold_4784 19968606 50 - 25762168 AUCUUGGCUAAUAGCCUUAAGUUAAUUGCCUUUUGGUUUUAGUCCGAUUU ...((((((((.((((..(((........)))..)))))))).))))... ( -11.70, z-score = -2.39, R) >droVir3.scaffold_12958 637552 50 + 3547706 AUUGCAAUUGCGAUAGUAUCGUUUCCUGUGCUGCUAUUUUAAUUCGAUCU ((((.(((((.(((((((.((.......)).))))))).))))))))).. ( -8.30, z-score = -0.97, R) >consensus AUCUUGGUUAAUAGCCUUAAGUUAAUUGCUUUUUGGUUUUAGUCCGAUUU ....(((((((.((((..((((.....))))...)))))))).))).... ( -4.57 = -4.47 + -0.11)

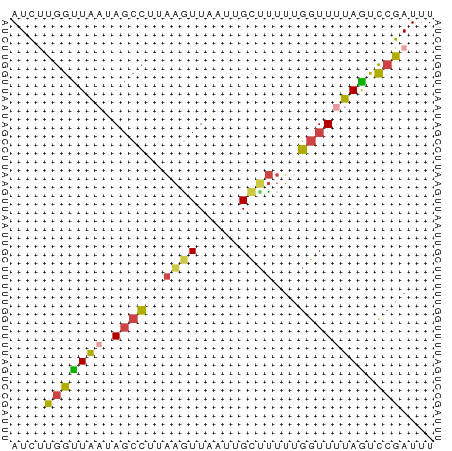
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:30 2011