
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,229,636 – 20,229,726 |
| Length | 90 |
| Max. P | 0.839301 |

| Location | 20,229,636 – 20,229,726 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.24 |
| Shannon entropy | 0.53950 |
| G+C content | 0.62944 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.03 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.839301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
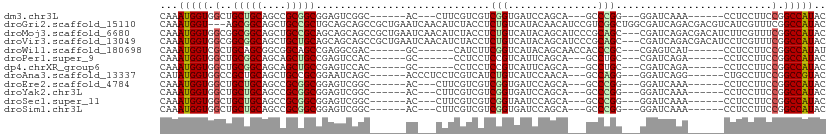
>dm3.chr3L 20229636 90 - 24543557 CAAAUGGUGGCUGCUGCAGCCGCGGCGGAGUCGGC------AC---CUUCGUCGUCGGUGAUCCAGCA---GCCCGG---GGAUCAAA------CCUCCUUCCGGCCAUAC ...(((((((((((((..((((((((((((.....------..---)))))))).))))....)))))---)))(((---(((.....------..)))..)))))))).. ( -43.40, z-score = -2.91, R) >droGri2.scaffold_15110 15388352 108 + 24565398 CAAAUGGU---AGCGGCAGCUGCCGCUGCAGCAGCCGCUGAAUCAACAUCUACCUCUGUCAUACAACAUCCGUCGGCUGGCGAUCAGACGACGUCAUCGUUUCGGCCAUAC ...(((((---((((((.(((((....))))).))))))(((.(............(((......)))..(((((.(((.....))).))))).....).))).))))).. ( -36.00, z-score = -1.63, R) >droMoj3.scaffold_6680 19273293 108 - 24764193 CAAAUGGUGGCGGCGGCAGCUGCCGCAGCAGCAGCCGCUGAAUCAACAUCUACCUCUGUCAUACAGCAUCCCGCAGC---CGAUCAGACGACAUCUUCGUUUCGGCCAUAC ...((((((((((((((.(((((....))))).)))))))...............(((.....)))....)))).((---(((...(((((.....))))))))))))).. ( -40.50, z-score = -2.85, R) >droVir3.scaffold_13049 18472453 108 - 25233164 CAAAUGGUGGCGGCGGCAGCUGCUGCAGCAGCAGCCGCUGAAUCAACAUCUACCUCUGUCAUACAGCAUCCCGCAGC---CGAUCAGACGACAUCCUCGUUUCGGCCAUAC ...((((((((((((((.(((((....))))).)))))))...............(((.....)))....)))).((---(((...(((((.....))))))))))))).. ( -41.20, z-score = -2.82, R) >droWil1.scaffold_180698 3124267 90 - 11422946 CAAAUGGUCGCUGCAGCGGCGGCAGCCGAGGCGAC------GC------CAUCUUCGGUCAUACAGCAACCACCCGC---CGAGUCAU------CCUCCUUCCGGCCAUAU ...(((((((((((....)))))....((((.(((------((------(......)))....(.((........))---.).)))..------)))).....)))))).. ( -27.40, z-score = -0.03, R) >droPer1.super_9 387734 87 - 3637205 CAAAUGGUGGCUGCGGCAGCAGCUGCCGAGUCCAC------GC------CCUCCUCCGUCAUUCAGCA---GCCUGC---CGAUCAGA------CCUCCUUCCGGCCAUAC ......(((((((((((((..(((((.((((..((------(.------.......))).)))).)))---))))))---))....((------......))))))))).. ( -31.10, z-score = -1.87, R) >dp4.chrXR_group6 2094506 87 - 13314419 CAAAUGGUGGCUGCGGCAGCAGCUGCCGAGUCCAC------GC------CCUCCUCCGUCAUUCAGCA---GCCUGC---CGAUCAGA------CCUCCUUCCGGCCAUAC ......(((((((((((((..(((((.((((..((------(.------.......))).)))).)))---))))))---))....((------......))))))))).. ( -31.10, z-score = -1.87, R) >droAna3.scaffold_13337 6352701 93 - 23293914 CAUAUGGUGGCCGCUGCAGCUGCCGCGGAAUCAGC------ACCCUCCUCGUCAUCUGUCAUCCAACA---GCCAGG---GGAUCAGG------CUGCCUUCCGGCCGUAC ..(((((((((.((....)).))).(((((.((((------....(((((.....((((......)))---)...))---)))....)------)))..))))))))))). ( -35.20, z-score = -1.09, R) >droEre2.scaffold_4784 19953962 90 - 25762168 CAAAUGGUGGCUGCUGCAGCCGCGGCGGAGUCGGC------AC---CUUCGUCGUCGGUGAUCCAGCA---GCCCGG---GGAUCAAA------CCUCCUUCCGGCCAUAC ...(((((((((((((..((((((((((((.....------..---)))))))).))))....)))))---)))(((---(((.....------..)))..)))))))).. ( -43.40, z-score = -2.91, R) >droYak2.chr3L 3854800 90 + 24197627 CAAAUGGUGGCUGCUGCAGCCGCGGCGGAGUCGGC------AC---CUUCGUCGUCGGUGAUCCAGCA---GCCCGG---GGAUCAAA------CCUCCUUCCGGCCAUAC ...(((((((((((((..((((((((((((.....------..---)))))))).))))....)))))---)))(((---(((.....------..)))..)))))))).. ( -43.40, z-score = -2.91, R) >droSec1.super_11 134229 90 - 2888827 CAAAUGGUGGCUGCUGCAGCCGCGGCGGAGUCGGC------AC---CUUCGUCGUCGGUAAUCCAGCA---GCCCGG---GGAUCAAA------CCUCCUUCCGGCCAUAC ...(((((((((((((..((((((((((((.....------..---)))))))).))))....)))))---)))(((---(((.....------..)))..)))))))).. ( -42.30, z-score = -3.12, R) >droSim1.chr3L 19600596 90 - 22553184 CAAAUGGUGGCUGCUGCAGCCGCGGCGGAGUCGGC------AC---CUUCGUCGUCGGUGAUCCAGCA---GCCCGG---GGAUCAAA------CCUCCUUCCGGCCAUAC ...(((((((((((((..((((((((((((.....------..---)))))))).))))....)))))---)))(((---(((.....------..)))..)))))))).. ( -43.40, z-score = -2.91, R) >consensus CAAAUGGUGGCUGCUGCAGCCGCGGCGGAGUCGGC______AC___CUUCGUCCUCGGUCAUCCAGCA___GCCCGC___CGAUCAAA______CCUCCUUCCGGCCAUAC ...(((((.(..(((((....))))).............................(((...............)))..........................).))))).. (-13.44 = -13.03 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:28 2011