
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,228,588 – 20,228,685 |
| Length | 97 |
| Max. P | 0.505437 |

| Location | 20,228,588 – 20,228,685 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.34 |
| Shannon entropy | 0.39879 |
| G+C content | 0.57482 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -17.66 |
| Energy contribution | -17.86 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.505437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20228588 97 + 24543557 -----------------GCGAAAAACUUACGUCUUCCUCCUGCGGAGUCCAUACAGGAUCCGUGUCAGAGUCGGUGAACUCA---UCCGGCUCGCCAACUACUCCACCAUCGCCGAC--- -----------------((((....................(((((.(((.....))))))))....(((((((((....))---.)))))))................))))....--- ( -28.00, z-score = -1.64, R) >droGri2.scaffold_15110 15387356 97 - 24565398 -----------------AAGUAACACUUACAUCUUCUUCCUGCGGUGUCCAGACGGGAUCCGUGUCCGAGUCGGUGAACUCC---UCCGGCUCACCCACCGCUCCGCCCUCCCCAAU--- -----------------..(((.....)))...........((((((........((((....))))(((((((.(.....)---.)))))))...))))))...............--- ( -26.60, z-score = -1.23, R) >droMoj3.scaffold_6680 19272284 97 + 24764193 -----------------AAGAUACACUUACAUCUUCCUCCUGCGGCGUCCAAACGGGAUCCGUGUCCGAGUCAGUGAACUCC---UCCGGCUCACCAACCGUGCCGCCCUCCCCAAU--- -----------------(((((........)))))......(((((((((.....))))..(((.(((((..((....)).)---).)))..))).......)))))..........--- ( -21.70, z-score = -0.39, R) >droVir3.scaffold_13049 18471447 97 + 25233164 -----------------AAGAAACACUUACAUCUUCUUCCUGCGGCGUCCAAACGGGAUCCGUGUCCGAGUCGGUGAACUCC---UCGGGCUCACCAACCGCACCGCCCUCCCCAAU--- -----------------.......................(((((.((((.....))))..(.(((((((..((....)).)---)))))).).....)))))..............--- ( -24.10, z-score = -0.20, R) >droWil1.scaffold_180698 3122799 97 + 11422946 -----------------AAAAGACACUUACAUCUUCUUCUUGCGGUGUCCAAACUGGAUCCGUAUCCGAAUCGGUAAACUCG---UCCGGCUCACCGACGGCAUUGCCCUCCCCACU--- -----------------...................(((.(((((.((((.....)))))))))...)))..(((((..(((---((.((....)))))))..))))).........--- ( -22.60, z-score = -1.07, R) >droPer1.super_9 386513 100 + 3637205 -----------------AAAUAACACUUACAUCAUCCUCUUGUGGAGUCCAGACAGGAUCCGUGUCCGAGUCGGUGAACUCGGCCUCCGGCUCGCCCACGGCACUGUUCUCCCCUGC--- -----------------..........................((((....(((((...(((((..((((((((.(........).))))))))..)))))..))))))))).....--- ( -33.80, z-score = -1.99, R) >dp4.chrXR_group6 2093285 100 + 13314419 -----------------AAAUAACACUUACAUCAUCCUCUUGUGGAGUCCAGACAGGAUCCGUGUCCGAGUCGGUGAACUCGGCCUCCGGCUCGCCCACGGCACUGUUCUCCCCUGC--- -----------------..........................((((....(((((...(((((..((((((((.(........).))))))))..)))))..))))))))).....--- ( -33.80, z-score = -1.99, R) >droAna3.scaffold_13337 6351497 117 + 23293914 GUGAGAUGAUGGUGUCUCUUCUGCACUUACGUCUUCCUCUUGCGGAGUCCAUACUGGAUCCGUGUCCGAGUCGGUGAACUCG---UCCUGCUCAUUAACGUUGUUACCAUCGCCAACUGC ((..(.((((((((........(((...((((.........(((((.(((.....))))))))....((((.((((....))---.)).))))....))))))))))))))))..))... ( -29.50, z-score = 0.11, R) >droEre2.scaffold_4784 19952934 97 + 25762168 -----------------GCGAAAAACUUACGUCUUCCUCCUGCGGAGUCCAAACAGGAUCCGUGUCCGAGUCGGUGAACUCA---UCCGGCUCGCCAACUACUCCACCAUCGCCGGC--- -----------------((((....................(((((.(((.....))))))))...((((((((((....))---.))))))))...............))))....--- ( -29.20, z-score = -1.68, R) >droYak2.chr3L 3853773 97 - 24197627 -----------------GCGAAACACUUACGUCUUCCUCCUGCGGAGUCCAUACAGGAUCCGUGUCCGAGUCGGUGAACUCA---UCCGGCUCGCCAACUACUCCACCAUCGCCGGC--- -----------------((((....................(((((.(((.....))))))))...((((((((((....))---.))))))))...............))))....--- ( -29.20, z-score = -1.46, R) >droSec1.super_11 133175 97 + 2888827 -----------------GCGAAAAACUUACGUCUUCCUCCUGCGGAGUCCAUACAGGAUCCGUGUCCGAGUCGGUGAACUCA---UCCGGCUCGCCAACUACUCCACCAUCGCCAGC--- -----------------((((....................(((((.(((.....))))))))...((((((((((....))---.))))))))...............))))....--- ( -29.20, z-score = -2.20, R) >droSim1.chr3L 19598899 97 + 22553184 -----------------GCGAAAAACUUACGUCUUCCUCCUGCGGAGUCCAUACAGGAUCCGUGUCCGAGUCGGUGAACUCA---UCCGGCUCGCCAACUACUCCACCAUCGCCGGC--- -----------------((((....................(((((.(((.....))))))))...((((((((((....))---.))))))))...............))))....--- ( -29.20, z-score = -1.60, R) >consensus _________________AAGAAACACUUACAUCUUCCUCCUGCGGAGUCCAUACAGGAUCCGUGUCCGAGUCGGUGAACUCA___UCCGGCUCGCCAACUGCUCCACCAUCCCCAGC___ .........................................((((.((((.....))))))))....(((((((............)))))))........................... (-17.66 = -17.86 + 0.19)

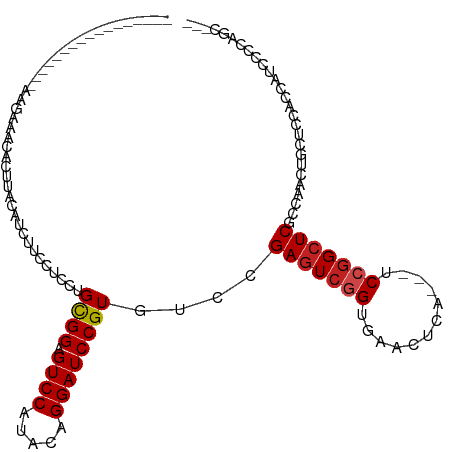
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:27 2011