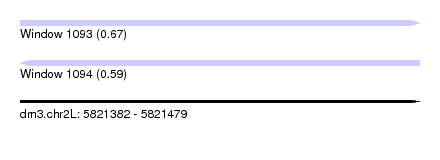
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,821,382 – 5,821,479 |
| Length | 97 |
| Max. P | 0.665749 |

| Location | 5,821,382 – 5,821,479 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 67.12 |
| Shannon entropy | 0.61900 |
| G+C content | 0.46698 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -9.50 |
| Energy contribution | -10.20 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.665749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5821382 97 + 23011544 -----UAGCGCAACUGGCGAGUGGCAUCGAAUCGAUUGUAUAAUUGUCGAUGCCAGUGGAUAUUUUGUUACGGCCCGAUUAGCCUCCAUUAAGCCAGUACCC- -----.......((((((....((((((((..(((((....)))))))))))))((((((...........(((.......)))))))))..))))))....- ( -32.30, z-score = -2.85, R) >droSim1.chr2L 5619401 99 + 22036055 ---UGCGAACUAACUGGCGACUGGCAUCAAAUCGAUUGUAUAAUUGUCGAUGCCAGUGGAUAUUUUGGUACGGCCCGAUUUGCCUCCAUUAAGCCAGUACCC- ---.........((((((.(((((((((....(((((....)))))..)))))))))(((((..((((......))))..))..))).....))))))....- ( -32.80, z-score = -3.01, R) >droSec1.super_5 3900267 99 + 5866729 ---UGCGAACUAACUGGCGACUGGCAUCAAAUCGAUUGUAUAAUUGUCGAUGCCAGUGGAUAUUUUGGUACGUCCCGCUUUGCCUCCAUUAAGCCAGUACCC- ---.........((((((.(((((((((....(((((....)))))..)))))))))(((......((((..........))))))).....))))))....- ( -30.20, z-score = -2.56, R) >droYak2.chr2L 8956373 102 - 22324452 UAGCAACUUGCAACUGCCGACUGGCAUCAAAUCGAUUGUAUAAUUCUCGAUGCCAGUGGAUAUUUAGGGUUUGCCCGGUUUCCCUCCAUUAAGCCAGUCCUC- ..((.....)).......(((((((.....(((((...........)))))...((((((......(((....)))((....))))))))..)))))))...- ( -29.10, z-score = -2.04, R) >droEre2.scaffold_4929 5914714 88 + 26641161 --------------UUGCAACUGGCAUCAAAUCGAUUGUAUAAUUGUCGAUGCCAGUGGAUAUUUAGGUUUCCACCGAUUUCCCUCCAUUAGGCCAGCUCUC- --------------..((.(((((((((....(((((....)))))..)))))))))(((.(((..(((....)))))).))).............))....- ( -22.20, z-score = -1.59, R) >dp4.chr4_group1 4760659 76 - 5278887 ---------------------UGGAAGCAAAUGGCUUGUAUAAUUGUCUUUGCCUGUGAAUAUUUCGGCGUUCUCC------CCAUCACCACCCCACUUUACG ---------------------(((..(((((.(((..........)))))))).............((.(....).------))....)))............ ( -9.20, z-score = 1.22, R) >droPer1.super_1 9767934 76 - 10282868 ---------------------UGGAAGCAAAUGGCUUGUAUAAUUGUCUUUGCCUGUGAAUAUUUCGGCGUUCUCC------CCAUCACCACCCCACUUUACG ---------------------(((..(((((.(((..........)))))))).............((.(....).------))....)))............ ( -9.20, z-score = 1.22, R) >consensus ___________AACUGGCGACUGGCAUCAAAUCGAUUGUAUAAUUGUCGAUGCCAGUGGAUAUUUUGGUAUGCCCCGAUUUGCCUCCAUUAAGCCAGUACCC_ .....................((((.....((((((.........))))))...((((((......((......))........))))))..))))....... ( -9.50 = -10.20 + 0.70)

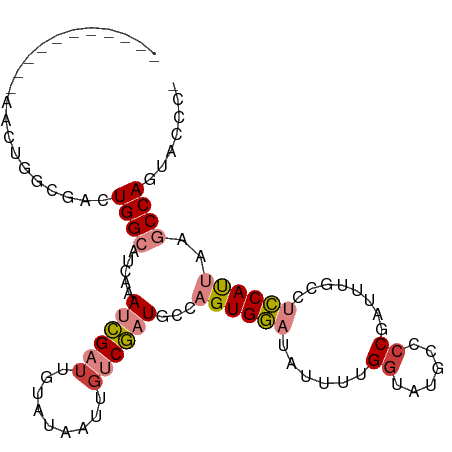
| Location | 5,821,382 – 5,821,479 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 67.12 |
| Shannon entropy | 0.61900 |
| G+C content | 0.46698 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -9.24 |
| Energy contribution | -10.00 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.588372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5821382 97 - 23011544 -GGGUACUGGCUUAAUGGAGGCUAAUCGGGCCGUAACAAAAUAUCCACUGGCAUCGACAAUUAUACAAUCGAUUCGAUGCCACUCGCCAGUUGCGCUA----- -(.((((((((....((((((((.....))))((......)).)))).(((((((((..(((........))))))))))))...))))).))).)..----- ( -32.00, z-score = -2.74, R) >droSim1.chr2L 5619401 99 - 22036055 -GGGUACUGGCUUAAUGGAGGCAAAUCGGGCCGUACCAAAAUAUCCACUGGCAUCGACAAUUAUACAAUCGAUUUGAUGCCAGUCGCCAGUUAGUUCGCA--- -(..(((((((....(((.(((.......)))...)))........(((((((((((..(((........)))))))))))))).))))))..)..)...--- ( -32.20, z-score = -2.48, R) >droSec1.super_5 3900267 99 - 5866729 -GGGUACUGGCUUAAUGGAGGCAAAGCGGGACGUACCAAAAUAUCCACUGGCAUCGACAAUUAUACAAUCGAUUUGAUGCCAGUCGCCAGUUAGUUCGCA--- -(..(((((((.....(((.((...))((......))......)))(((((((((((..(((........)))))))))))))).))))))..)..)...--- ( -30.70, z-score = -2.16, R) >droYak2.chr2L 8956373 102 + 22324452 -GAGGACUGGCUUAAUGGAGGGAAACCGGGCAAACCCUAAAUAUCCACUGGCAUCGAGAAUUAUACAAUCGAUUUGAUGCCAGUCGGCAGUUGCAAGUUGCUA -...(((((.(.....(((((....))(((....)))......)))((((((((((((..............)))))))))))).).)))))((.....)).. ( -34.94, z-score = -2.94, R) >droEre2.scaffold_4929 5914714 88 - 26641161 -GAGAGCUGGCCUAAUGGAGGGAAAUCGGUGGAAACCUAAAUAUCCACUGGCAUCGACAAUUAUACAAUCGAUUUGAUGCCAGUUGCAA-------------- -((...((..((....))..))...))(((....))).........(((((((((((..(((........)))))))))))))).....-------------- ( -24.20, z-score = -1.70, R) >dp4.chr4_group1 4760659 76 + 5278887 CGUAAAGUGGGGUGGUGAUGG------GGAGAACGCCGAAAUAUUCACAGGCAAAGACAAUUAUACAAGCCAUUUGCUUCCA--------------------- .((((((((((.(((((....------......))))).....))))).(((................))).))))).....--------------------- ( -14.09, z-score = 0.74, R) >droPer1.super_1 9767934 76 + 10282868 CGUAAAGUGGGGUGGUGAUGG------GGAGAACGCCGAAAUAUUCACAGGCAAAGACAAUUAUACAAGCCAUUUGCUUCCA--------------------- .((((((((((.(((((....------......))))).....))))).(((................))).))))).....--------------------- ( -14.09, z-score = 0.74, R) >consensus _GGGAACUGGCUUAAUGGAGGCAAAUCGGGCAAUACCAAAAUAUCCACUGGCAUCGACAAUUAUACAAUCGAUUUGAUGCCAGUCGCCAGUU___________ ...............((((........((......))......))))..((((((((................))))))))...................... ( -9.24 = -10.00 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:54 2011