
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,191,553 – 20,191,626 |
| Length | 73 |
| Max. P | 0.988166 |

| Location | 20,191,553 – 20,191,626 |
|---|---|
| Length | 73 |
| Sequences | 4 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 63.54 |
| Shannon entropy | 0.58141 |
| G+C content | 0.42158 |
| Mean single sequence MFE | -19.20 |
| Consensus MFE | -8.83 |
| Energy contribution | -10.21 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20191553 73 + 24543557 CAUCAGCUGAUACUC-GUGCUGCAG-GCAGAGUAUCAAAAAUAAGCUAUU----AACAGCUAUUAAAGGGUAUCAGUAG----- .....((((((((((-...(((...-.))).............((((...----...))))......))))))))))..----- ( -20.40, z-score = -2.95, R) >droEre2.scaffold_4784 19920693 84 + 25762168 CGCCUGCUAAAACUCAGUGGUAAAUGAAGGGGUACGGAAUUUAAUUUAAUUUACAAGAGCUGCUAAAGGGUAUCAGAGGUGUUA (((((.((.(.((((..(((((......(.....).(((((......)))))........)))))..)))).).)))))))... ( -13.50, z-score = 0.94, R) >droSec1.super_11 101754 75 + 2888827 CAUCAGCUGAUACUC-GACUCGAGCUGCACGGUAUUAAAACUCAGUAC------AACAGGUAUUAAAGGGUAUCAGCAGUGG-- .....((((((((((-......(.(((....(((((.......)))))------..))).)......)))))))))).....-- ( -22.10, z-score = -3.23, R) >droSim1.chr3L 19479097 70 + 22553184 CAUCAGCUGAUACUC-GAGCUGCAG-G----GUAUCAAAACGCAGUAA------AACAGCUAUUAAAGGGUAUCAGCAGUAG-- .....((((((((((-.(((((...-.----((........)).....------..)))))......)))))))))).....-- ( -20.80, z-score = -3.70, R) >consensus CAUCAGCUGAUACUC_GAGCUGAAG_GCAGGGUAUCAAAACUAAGUAA______AACAGCUAUUAAAGGGUAUCAGCAGUGG__ .....((((((((((..(((((..................................)))))......))))))))))....... ( -8.83 = -10.21 + 1.38)
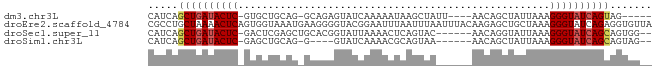
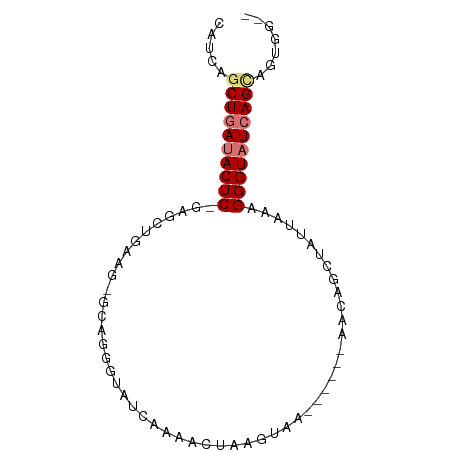
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:25 2011