
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,189,477 – 20,189,594 |
| Length | 117 |
| Max. P | 0.982622 |

| Location | 20,189,477 – 20,189,571 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.76 |
| Shannon entropy | 0.42846 |
| G+C content | 0.43494 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -16.94 |
| Energy contribution | -17.45 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.964214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
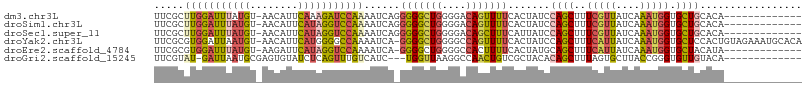
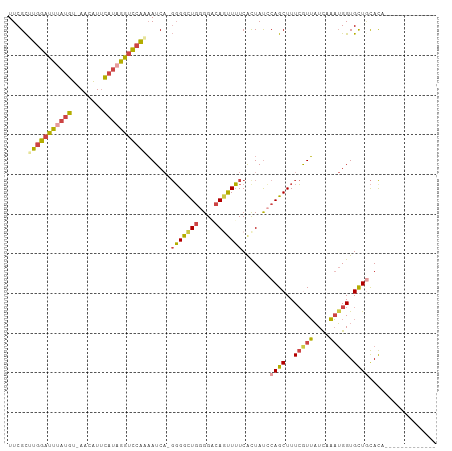
>dm3.chr3L 20189477 94 - 24543557 UUCGCUUGGAUUUAUGU-AACAUUCAAAGAUCCAAAAUCAGGGGGCUGGGGACAGUUUUCACUAUCCAGCUUUCGUUAUCAAAUGGUGCUGCACA------------- .....((((((((.((.-......)).)))))))).....(..(((((....)))))..)......((((..(((((....))))).))))....------------- ( -26.90, z-score = -2.38, R) >droSim1.chr3L 19477041 94 - 22553184 UUCGCUUGGAUUUAUGU-AACAUUCAUAGGUCCAAAAUCAGGGGGCUGGGGACAGUUUUCACUAUCCAGCUUUCGUUAUCAAAUGGUGCUGCACA------------- .....(((((((((((.-......))))))))))).....(..(((((....)))))..)......((((..(((((....))))).))))....------------- ( -30.50, z-score = -3.51, R) >droSec1.super_11 99690 94 - 2888827 UUCGCUUGGAUUUAUGU-AACAUUCAUAGGUCCAAAAUCAGGGGGCUGGGGACAGCUUUCAUUAUCCAGCUUUCGUUAUCAAAUGGUGCUGCACA------------- .....(((((((((((.-......))))))))))).....(..(((((....)))))..)......((((..(((((....))))).))))....------------- ( -33.00, z-score = -4.23, R) >droYak2.chr3L 3822988 106 + 24197627 UUCGCGUGGAUUAAUGU-AACAUUCAUGGGGCCAAAAUCA-GGGGCUGGGGCCAGUUUUCACUAUCCAGCUUUCAUUAUCAAAUGGUGCUCCACUGUAGAAAUGCACA ...((((......))))-........(((((((.......-(((((((((...(((....))).)))))))))((((....))))).)))))).((((....)))).. ( -25.90, z-score = 0.26, R) >droEre2.scaffold_4784 19918377 93 - 25762168 UUCGCGUGGAUUUAUGU-AAGAUUCAUAGGUCCAAAAUCA-GGGGCUGGGGCCACUUUUCACUAUGCAGCUUUCAUUAUCAAAUGGUGCUACAUA------------- ...(((((((((((((.-......)))))))))......(-(((((....))).)))......))))(((..(((((....))))).))).....------------- ( -23.50, z-score = -1.27, R) >droGri2.scaffold_15245 14587686 91 - 18325388 UUCGUAU-GAUUAAUGCGAGUGUAUCUCAGUUUGUCAUC---UGGUUAAGGCCAACUGUCGCUACACAGCUUUAGUGCUUACCGGGUGUUGUACA------------- (((((((-.....))))))).........((....((((---((((..((((..((((..(((....)))..))))))))))))))))....)).------------- ( -25.90, z-score = -1.41, R) >consensus UUCGCUUGGAUUUAUGU_AACAUUCAUAGGUCCAAAAUCA_GGGGCUGGGGACAGUUUUCACUAUCCAGCUUUCGUUAUCAAAUGGUGCUGCACA_____________ .....(((((((((((........)))))))))))......(((((((....))))))).......((((..(((((....))))).))))................. (-16.94 = -17.45 + 0.51)

| Location | 20,189,502 – 20,189,594 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 74.84 |
| Shannon entropy | 0.45120 |
| G+C content | 0.50002 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -17.19 |
| Energy contribution | -17.87 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
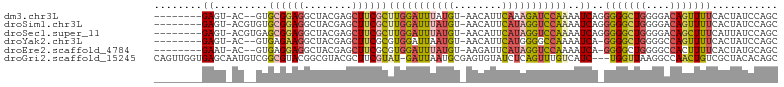
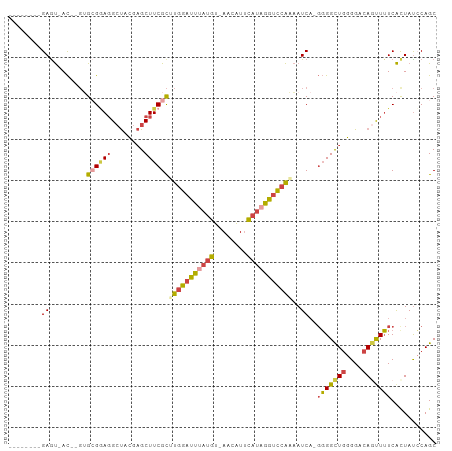
>dm3.chr3L 20189502 92 - 24543557 --------GAGU-AC--GUGCGGAGGCUACGAGCUUCGCUUGGAUUUAUGU-AACAUUCAAAGAUCCAAAAUCAGGGGGCUGGGGACAGUUUUCACUAUCCAGC --------.(((-..--..(((.((((.....)))))))((((((((.((.-......)).)))))))).....(..(((((....)))))..))))....... ( -28.70, z-score = -2.31, R) >droSim1.chr3L 19477066 94 - 22553184 --------GAGU-ACGUGUGCGGAGGCUACGAGCUUCGCUUGGAUUUAUGU-AACAUUCAUAGGUCCAAAAUCAGGGGGCUGGGGACAGUUUUCACUAUCCAGC --------.(((-......(((.((((.....)))))))(((((((((((.-......))))))))))).....(..(((((....)))))..))))....... ( -32.30, z-score = -2.81, R) >droSec1.super_11 99715 94 - 2888827 --------GAGU-ACGUGAGCGGAGGCUACGAGCUUCGCUUGGAUUUAUGU-AACAUUCAUAGGUCCAAAAUCAGGGGGCUGGGGACAGCUUUCAUUAUCCAGC --------....-...((((((.((((.....)))))))(((((((((((.-......)))))))))))..)))(..(((((....)))))..).......... ( -35.60, z-score = -4.03, R) >droYak2.chr3L 3823026 91 + 24197627 --------GAGU-AC--GUGAGAAGGCUACGAGCUUCGCGUGGAUUAAUGU-AACAUUCAUGGGGCCAAAAUCA-GGGGCUGGGGCCAGUUUUCACUAUCCAGC --------(..(-(.--(((((((((((.(.((((((...(((....(((.-......)))....)))......-)))))).)))))..)))))))))..)... ( -26.20, z-score = -0.72, R) >droEre2.scaffold_4784 19918402 91 - 25762168 --------GAAU-AC--GUGAGGAGGCUACGAGCUUCGCGUGGAUUUAUGU-AAGAUUCAUAGGUCCAAAAUCA-GGGGCUGGGGCCACUUUUCACUAUGCAGC --------....-..--(((((((((((.(.((((((...((((((((((.-......))))))))))......-)))))).)))))..)))))))........ ( -32.40, z-score = -3.26, R) >droGri2.scaffold_15245 14587711 100 - 18325388 CAGUUGGUGAGCAAUGUCGGCGUACGGCGUACGCUUCGUAU-GAUUAAUGCGAGUGUAUCUCAGUUUGUCAUC---UGGUUAAGGCCAACUGUCGCUACACAGC ((((((((.(((...(..((((.((((.(((((((.(((((-.....)))))))))))).)).)).))))..)---..)))...))))))))..(((....))) ( -32.50, z-score = -1.28, R) >consensus ________GAGU_AC__GUGCGGAGGCUACGAGCUUCGCUUGGAUUUAUGU_AACAUUCAUAGGUCCAAAAUCA_GGGGCUGGGGACAGUUUUCACUAUCCAGC ...................((((((........))))))(((((((((((........)))))))))))......(((((((....)))))))........... (-17.19 = -17.87 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:24 2011