
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,803,458 – 5,803,559 |
| Length | 101 |
| Max. P | 0.508739 |

| Location | 5,803,458 – 5,803,559 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 58.11 |
| Shannon entropy | 0.76260 |
| G+C content | 0.66793 |
| Mean single sequence MFE | -36.96 |
| Consensus MFE | -14.66 |
| Energy contribution | -16.36 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
| WARNING | Out of training range. z-scores are NOT reliable. |
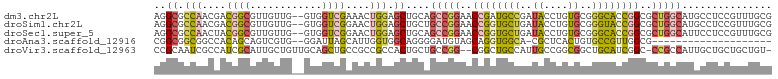
Download alignment: ClustalW | MAF
>dm3.chr2L 5803458 101 + 23011544 CGCAAACGGAGGCAUGCCAGCGCCGGUGCCCGCACAGGUAUCGGCAUCGGUUCCGGCUGCAGCUCCAGUUUCGACCAC--CAACAACGCCGUCGUUGGCGCCU ((.((((((((.(.(((.((((((((((((......)))))))))..((....))))))))))))).)))))).....--......(((((....)))))... ( -41.20, z-score = -1.00, R) >droSim1.chr2L 5602453 101 + 22036055 CGCAAACGGAGGCAUGCCAGCGCCGGUACCCGCACAGGUAUCAGCACCGGUUCCGGCAGCAGCUCCAGUUCCGACCAC--CAACAACGCCGUCGUUGGCGCCU (((.((((..(((..(((.(.((.((((((......)))))).)).).)))..(((.(((.......)))))).....--.......)))..)))).)))... ( -34.30, z-score = -0.29, R) >droSec1.super_5 3882549 101 + 5866729 CGCAAACGGAGGAAUGCCAGCGCCGGUGCCCGCACAGGUAUCAGCACCGGUUCCGGCUGCAGCUCCAGUUCCGACCAC--CAACAACGCCGUAGUUGGCGCCU .((...(((((((.(((.(((((((((((((.....)).....))))))))....))))))..)))...)))).....--.......((((....)))))).. ( -37.40, z-score = -0.61, R) >droAna3.scaffold_12916 11225429 80 + 16180835 --------------------CGCCAACGGCACAGUGAGCG-UGCCACCUGCUACAUCCCCUGCCACCAAUGCUAAUCC--CACGACUGCUGUGGCCGCCGCCG --------------------.(((...)))...(((.(((-.(((((..((..........((.......))......--.......)).))))))))))).. ( -20.60, z-score = 0.01, R) >droVir3.scaffold_12963 15910186 99 - 20206255 -ACAGCAGCAGCAAUGGCGG-GCCGAUGCAGCCGCCGGCAAUGGCAGCCG--CCGGCAGCAGUGGCGGCGGCAGCUGCAACAGCAAUGCGAUGGCGAUUGCGG -...(((((.(((.(((...-.))).))).(((((((.((...((.(((.--..))).))..)).))))))).)))))....(((((((....)).))))).. ( -51.30, z-score = -1.39, R) >consensus CGCAAACGGAGGAAUGCCAGCGCCGGUGCCCGCACAGGUAUCGGCACCGGUUCCGGCAGCAGCUCCAGUUCCGACCAC__CAACAACGCCGUCGUUGGCGCCU ..........((....))...(((((((((......)))))))))...(((.(((((.((.((........................)).)).))))).))). (-14.66 = -16.36 + 1.70)

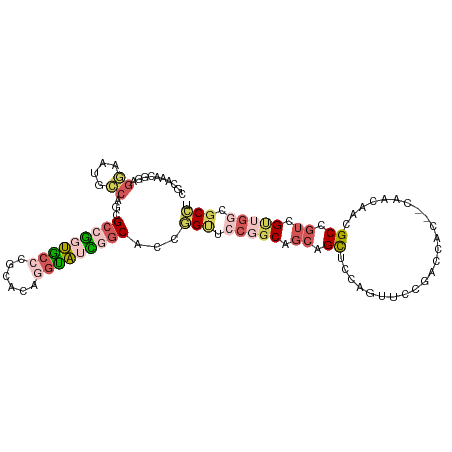

| Location | 5,803,458 – 5,803,559 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 58.11 |
| Shannon entropy | 0.76260 |
| G+C content | 0.66793 |
| Mean single sequence MFE | -42.70 |
| Consensus MFE | -16.94 |
| Energy contribution | -18.38 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.508739 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 5803458 101 - 23011544 AGGCGCCAACGACGGCGUUGUUG--GUGGUCGAAACUGGAGCUGCAGCCGGAACCGAUGCCGAUACCUGUGCGGGCACCGGCGCUGGCAUGCCUCCGUUUGCG .((((((((((((...)))))))--)).))).((((.((((..((((((((..(((.((((..((....))..)))).)))..))))).)))))))))))... ( -44.80, z-score = -0.73, R) >droSim1.chr2L 5602453 101 - 22036055 AGGCGCCAACGACGGCGUUGUUG--GUGGUCGGAACUGGAGCUGCUGCCGGAACCGGUGCUGAUACCUGUGCGGGUACCGGCGCUGGCAUGCCUCCGUUUGCG .((((((((((((...)))))))--)).))).((((.((((..((((((((..((((((((..((....))..))))))))..)))))).))))))))))... ( -48.20, z-score = -1.65, R) >droSec1.super_5 3882549 101 - 5866729 AGGCGCCAACUACGGCGUUGUUG--GUGGUCGGAACUGGAGCUGCAGCCGGAACCGGUGCUGAUACCUGUGCGGGCACCGGCGCUGGCAUUCCUCCGUUUGCG .((((((......))))))....--(..(.((((...((((.....(((((..((((((((..((....))..))))))))..))))).)))))))).)..). ( -43.30, z-score = -0.33, R) >droAna3.scaffold_12916 11225429 80 - 16180835 CGGCGGCGGCCACAGCAGUCGUG--GGAUUAGCAUUGGUGGCAGGGGAUGUAGCAGGUGGCA-CGCUCACUGUGCCGUUGGCG-------------------- (((((((.((((..((((((...--.)))).))..)))).(((((((.(((.((.....)))-))))).)))))))))))...-------------------- ( -30.40, z-score = -0.08, R) >droVir3.scaffold_12963 15910186 99 + 20206255 CCGCAAUCGCCAUCGCAUUGCUGUUGCAGCUGCCGCCGCCACUGCUGCCGG--CGGCUGCCAUUGCCGGCGGCUGCAUCGGC-CCGCCAUUGCUGCUGCUGU- ..(((((.((....)))))))....(((((((((((.((....)).).)))--)))))))......(((((((.(((..((.-...))..))).))))))).- ( -46.80, z-score = -0.56, R) >consensus AGGCGCCAACCACGGCGUUGUUG__GUGGUCGCAACUGGAGCUGCAGCCGGAACCGGUGCCGAUACCUGUGCGGGCACCGGCGCUGGCAUGCCUCCGUUUGCG ..((.(((....((((............))))....))).))....(((((..((((((((..((....))..))))))))..)))))............... (-16.94 = -18.38 + 1.44)
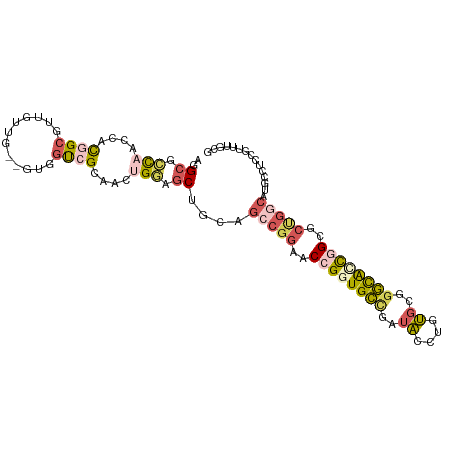
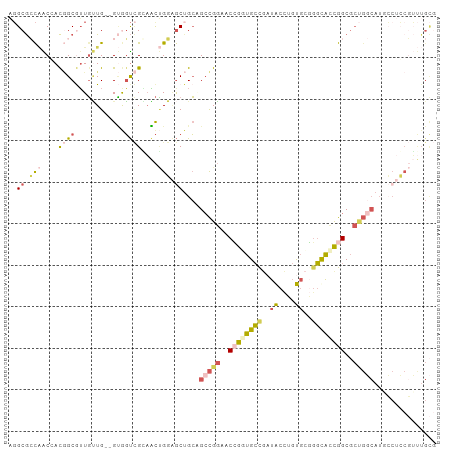
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:53 2011