
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,145,303 – 20,145,404 |
| Length | 101 |
| Max. P | 0.627360 |

| Location | 20,145,303 – 20,145,404 |
|---|---|
| Length | 101 |
| Sequences | 14 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.47 |
| Shannon entropy | 0.43934 |
| G+C content | 0.58385 |
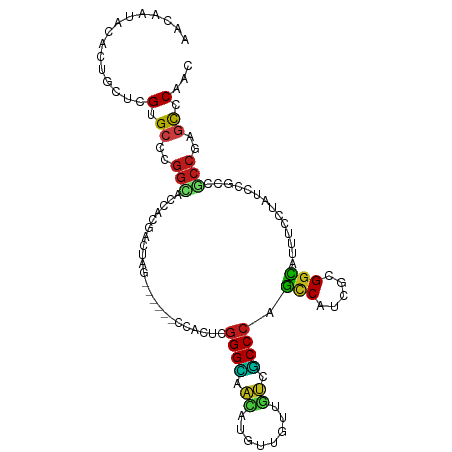
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -15.44 |
| Energy contribution | -14.84 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.80 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
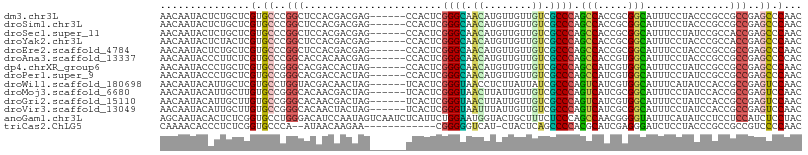
>dm3.chr3L 20145303 101 - 24543557 AACAAUACUCUGCUCGUGCCCGGCUCCACGACGAG------CCACUCGGGCAACAUGUUGUUGUCGCCCAGCCACCGCGGCAUUUCCUACCCGCCGCCGAGCCCAAC ...........(((((.((..(((((......)))------))....((((.(((......))).)))).))....(((((...........))))))))))..... ( -36.10, z-score = -2.89, R) >droSim1.chr3L 19433082 101 - 22553184 AACAAUACUCUGCUCGUGCCCGGCUCCACGACGAG------CCACUCGGGCAACAUGUUGUUGUCGCCCAGCCACCGCGGCAUUUCCUACCCGCCGCCGAGCCCAAC ...........(((((.((..(((((......)))------))....((((.(((......))).)))).))....(((((...........))))))))))..... ( -36.10, z-score = -2.89, R) >droSec1.super_11 55915 101 - 2888827 AACAAUACUCUGCUCGUGCCCGGCUCCACGACGAG------CCACUCGGGCAACAUGUUGUUGUCGCCCAGCCACCGCGGCAUUUCCUAUCCGCCACCGAGCCCAAC ...........(((((.((..(((((......)))------))....((((.(((......))).)))).))....((((.((.....))))))...)))))..... ( -32.70, z-score = -2.31, R) >droYak2.chr3L 3774142 101 + 24197627 AACAAUACUCUACUCGUGCCCGGCUCCACGACGAG------CCACUCGGGCAACAUGUUGUUGUCGCCCAGCCACCGCGGCAUUUCCUACCCGCCACCGAGCCCAAC ............((((.((..(((((......)))------))....((((.(((......))).)))).......))(((...........)))..))))...... ( -28.90, z-score = -1.63, R) >droEre2.scaffold_4784 19872587 101 - 25762168 AACAAUACUCUGCUCGUGCCCGGCUCCACGACGAG------CCACUCGGGCAACAUGUUGUUGUCGCCCAGCCACCGCGGCAUUUCCUACCCGCCGCCGAGCCCAAC ...........(((((.((..(((((......)))------))....((((.(((......))).)))).))....(((((...........))))))))))..... ( -36.10, z-score = -2.89, R) >droAna3.scaffold_13337 6267860 101 - 23293914 AACAAUACCCUUCUCGUGCCCGGCACCACAACGAG------CCACUCGGGCAACAUGUUGUUGUCGCCCAGCCACCGUGGCAUUUCCUACCCGCCGCCGAGCCCCAC ............((((.....(((........(((------...)))((((.(((......))).)))).)))...(((((...........)))))))))...... ( -28.00, z-score = -1.19, R) >dp4.chrXR_group6 2019517 101 - 13314419 AACAAUACCCUGCUCGUGCCGGGCACGACCACUAG------CCACUCGGGCAACAUGUUGUUGUCGCCCAGCCAUCGUGGCAUUUCCUAUCCGCCGCCGAGCCCAAC ...........(((((.((.(((((((((.((..(------((.....))).....)).))))).)))).))....(((((...........))))))))))..... ( -33.80, z-score = -1.89, R) >droPer1.super_9 312704 101 - 3637205 AACAAUACCCUGCUCGUGCCGGGCACGACCACUAG------CCACUCGGGCAACAUGUUGUUGUCGCCCAGCCAUCGUGGCAUUUCCUAUCCGCCGCCGAGCCCAAC ...........(((((.((.(((((((((.((..(------((.....))).....)).))))).)))).))....(((((...........))))))))))..... ( -33.80, z-score = -1.89, R) >droWil1.scaffold_180698 8206419 101 + 11422946 AACAAUACAUUGCUCGUGCCUGGUACGACAACUAG------UCACUCGGGUAACCUCUUAUUAUCGCCCAGUCAUCGUGGCAUUUCAUAUCCACCGCCGAGUCCAAC ...........(((((.((((((...(((.....)------))....((....))............)))).....((((.((.....)))))).)))))))..... ( -21.00, z-score = -0.74, R) >droMoj3.scaffold_6680 19301534 101 - 24764193 AACAAUACAUUGCUUGUGCCGGGCACAACGACUAG------UCACUCGGGUAACUUAUUGUUGUCGCCCAGUCAUCGCGGCAUUUCCUAUCCACCGCCGAGUCCAAC ...........(((((.((.(((((((((((..((------(.((....)).)))..))))))).)))).(((.....)))..............)))))))..... ( -28.00, z-score = -1.56, R) >droGri2.scaffold_15110 15416827 101 + 24565398 AACAAUACAUUGCUUGUGCCGGGCACAACGACUAG------UCACUCGGGUAACUUAUUGUUGUCGCCCAGUCAUCGUGGCAUUUCCUAUCCACCGCCGAGUCCAAC ...........(((((.((.(((((((((((..((------(.((....)).)))..))))))).)))).......((((.((.....)))))).)))))))..... ( -29.80, z-score = -2.11, R) >droVir3.scaffold_13049 18502039 101 - 25233164 AACAAUACAUUGCUUGUGCCGGGCACAACUACUAG------UCACUCGGGUAAUUUAUUGUUGUCGCCCAGUCAUCGCGGCAUUUCCUAUCCACCGCCGAGUCCAAC ...........(((((.((.(((((((((....((------(.((....)).)))....))))).)))).(((.....)))..............)))))))..... ( -23.10, z-score = -0.44, R) >anoGam1.chr3L 313098 107 - 41284009 AGCAAUACACUCUCGGUGCCUGGGACAUCCAAUAGUCAAUCUCAUUCUGGAAUGGUACUGCUUUCUCCCAGCCAACGGGGUAUUUCAUAUCCUCCUCCAUCUCCUAC .(.(((((...((((.((.((((((....((.((.(((.(((......))).))))).)).....)))))).)).))))))))).)..................... ( -19.60, z-score = 0.73, R) >triCas2.ChLG5 3368946 92 + 18847211 CAAAACACCCUCUCGGUGCCCA--AUAACAAGAA------------CGGGGGUCAU-CUACUCAGCCCCACGCAUCGAGGGAUCUCCUACCCGCCGCCGUCCCCAAC .........((((((((((...--....(.....------------.)(((((...-.......)))))..)))))))))).......................... ( -22.50, z-score = -0.45, R) >consensus AACAAUACACUGCUCGUGCCCGGCACCACGACUAG______CCACUCGGGCAACAUGUUGUUGUCGCCCAGCCAUCGCGGCAUUUCCUAUCCGCCGCCGAGCCCAAC ...............(.((..(((.......................((((.((........)).)))).(((.....)))..............)))..)).)... (-15.44 = -14.84 + -0.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:14 2011