
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,141,888 – 20,141,980 |
| Length | 92 |
| Max. P | 0.754084 |

| Location | 20,141,888 – 20,141,980 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 82.01 |
| Shannon entropy | 0.34842 |
| G+C content | 0.57658 |
| Mean single sequence MFE | -18.90 |
| Consensus MFE | -16.65 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.754084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20141888 92 - 24543557 CACUUAAAACAGGCGAUGCAUGCAGCAGCCAAGAGGAUUAUUUGCAUGCCACGCCCCC-AGUCCCCAGUCCCCAUCCACUCA-CCGCCCAUCGC ...........((((..((((((((...((....)).....))))))))..))))...-.......................-........... ( -19.20, z-score = -1.11, R) >droSim1.chr3L 19429676 93 - 22553184 CACUUUAAACAGGCGAUGCAUGCAGCAGCCAAGAGGAUUAUUUGCAUGCCACGCCCCCUGGUCCCUAUUCACCAACCACCCA-CCGCCCACCGC ...........((((..((((((((...((....)).....))))))))..))))...((((........))))........-........... ( -21.70, z-score = -1.39, R) >droSec1.super_11 52534 89 - 2888827 CACUUCAACCAGGCGAUGCAUGCAGCAGCCAAGAGGAUUAUUUGCAUGCCACGUCCCC-AGUCCCUGCUCACCAACCACCCA-CCGCCCAC--- ...........((((..((((((((...((....)).....))))))))........(-((...)))...............-.))))...--- ( -18.00, z-score = -0.59, R) >droYak2.chr3L 3768234 93 + 24197627 CACUUAAAACAGACGAUGCAUGCAGCUGCCAAGAGGAUUGUUUGCAUGCCACGCCCCC-AGUCCGUAUUCCCCAUCCACGCAUCCACCCAACGC ..............(((((..((....)).....((((((...((.......))...)-)))))...............))))).......... ( -16.60, z-score = -0.16, R) >droEre2.scaffold_4784 19869182 91 - 25762168 CACUUAAACCAGGCGAUGCAUGCAGCAGCCAAGAGCAUUAUUUGCGUGCCACGCCCCC-AGUCCGUGUUCCCCAUCCACCC--CCGCCCCUCGC ...........((((..((((((((..((.....)).....))))))))((((.....-....))))..............--.))))...... ( -18.40, z-score = -0.84, R) >droAna3.scaffold_13337 6264487 92 - 23293914 CCUUUCACACAGGCGAUGCAUGCAGCAGCCAAGUGGAUUAUUUGCAUGCCACGCCCCA-UAUCCACAACAUUCAACCUCUAU-CCGCCCACUUA ...........((((..((((((((...((....)).....))))))))..))))...-.......................-........... ( -19.50, z-score = -1.74, R) >consensus CACUUAAAACAGGCGAUGCAUGCAGCAGCCAAGAGGAUUAUUUGCAUGCCACGCCCCC_AGUCCCUAUUCCCCAACCACCCA_CCGCCCACCGC ...........((((..((((((((...((....)).....))))))))..))))....................................... (-16.65 = -16.73 + 0.09)

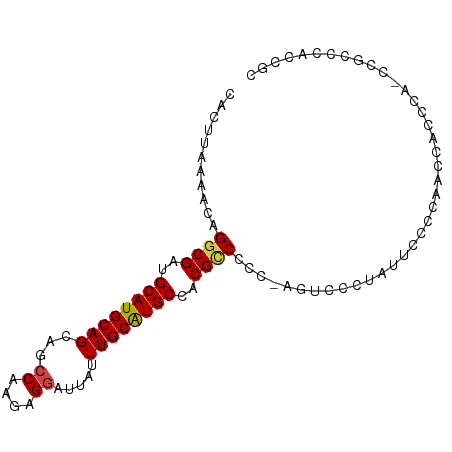
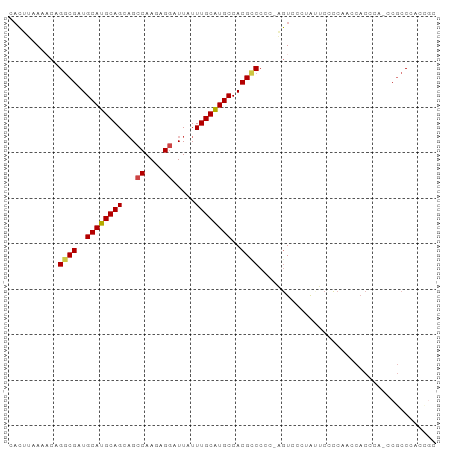
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:13 2011