
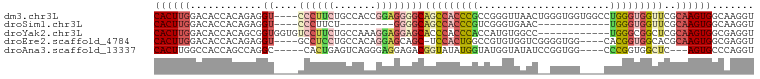
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,139,424 – 20,139,520 |
| Length | 96 |
| Max. P | 0.774251 |

| Location | 20,139,424 – 20,139,520 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 62.29 |
| Shannon entropy | 0.65297 |
| G+C content | 0.64292 |
| Mean single sequence MFE | -41.22 |
| Consensus MFE | -15.13 |
| Energy contribution | -17.01 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20139424 96 + 24543557 CACUUGGACACCACAGAGGU----CCCUUCUGCCACCGGAGGGGCAGCCACCCGCCGGGUUAACUGGGUGGUGGCCUGGGUGGUUCGCAAGUGGCAAGGU ((((((((.(((((..((((----((((((.......)))))))).((((((.(((.((....)).)))))))))))..))))))).))))))....... ( -49.00, z-score = -1.93, R) >droSim1.chr3L 19427127 75 + 22553184 CACUUGGACACCACAGAGGU----CCCUUCU---------GGGGCAGCCACCCGUCGGGUGAAC------------UGGGUGGUUCGCAAGUGGCAAGGU ((((((((.(((((..((((----(((....---------)))))...(((((...)))))..)------------)..))))))).))))))....... ( -33.70, z-score = -1.63, R) >droYak2.chr3L 3766041 88 - 24197627 CACUUGGACACCACAGCGGUGGUGUCCUUCUGCCAAAGGAGGAGCACCCACCCACCAUGUGGCC------------UGGGCGGCUCGCAAGUGGCGAGGU ..((((((((((((....)))))))))....((((...(..((((.((((.((((...))))..------------))))..)))).)...))))))).. ( -40.70, z-score = -1.46, R) >droEre2.scaffold_4784 19867074 91 + 25762168 CACUUGGACACCACAGAGGU----GCCUCCUGCCACAGGAGCAGC-UCCACUGGCCGUGUGGUCGGGGUGG----CACGGUGGCACGCAAGUGGCGAGGU ..((((....((((....((----(((.((((((((.((((...)-))).((((((....)))))).))))----)).)).)))))....)))))))).. ( -43.50, z-score = -1.05, R) >droAna3.scaffold_13337 6262242 88 + 23293914 CACUUGGCCACCAGCCAGGC-----CACUGAGUCAGGGAGGAGACGGUAUAUGGUAUGGUAUAUCCGGUGG----CCCGGUGGCUC---AGUGCCCAGGU .((((((.(((.((((((((-----(((((.(((........)))(((((((......)))))))))))))----))...))))).---.))).)))))) ( -39.20, z-score = -1.60, R) >consensus CACUUGGACACCACAGAGGU____CCCUUCUGCCACAGGAGGGGCAGCCACCCGCCGGGUGAAC__GGUGG____CUGGGUGGCUCGCAAGUGGCAAGGU ..((((..(((.............((((((.......))))))(((((((((((......................))))))))).))..))).)))).. (-15.13 = -17.01 + 1.88)


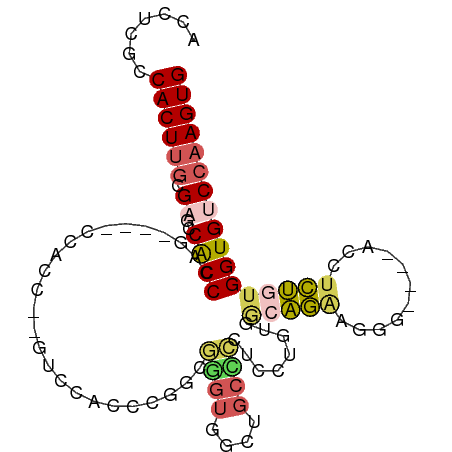
| Location | 20,139,424 – 20,139,520 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 62.29 |
| Shannon entropy | 0.65297 |
| G+C content | 0.64292 |
| Mean single sequence MFE | -36.84 |
| Consensus MFE | -14.18 |
| Energy contribution | -14.30 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20139424 96 - 24543557 ACCUUGCCACUUGCGAACCACCCAGGCCACCACCCAGUUAACCCGGCGGGUGGCUGCCCCUCCGGUGGCAGAAGGG----ACCUCUGUGGUGUCCAAGUG .......((((((.(((((((..(((((.((((((.(((.....)))))))))(((((((...)).)))))...))----.)))..))))).)))))))) ( -42.50, z-score = -1.64, R) >droSim1.chr3L 19427127 75 - 22553184 ACCUUGCCACUUGCGAACCACCCA------------GUUCACCCGACGGGUGGCUGCCCC---------AGAAGGG----ACCUCUGUGGUGUCCAAGUG .......((((((.(((((((.((------------((.(((((...))))))))).(((---------....)))----......))))).)))))))) ( -28.90, z-score = -1.50, R) >droYak2.chr3L 3766041 88 + 24197627 ACCUCGCCACUUGCGAGCCGCCCA------------GGCCACAUGGUGGGUGGGUGCUCCUCCUUUGGCAGAAGGACACCACCGCUGUGGUGUCCAAGUG ..(((((.....)))))((((((.------------.(((....))))))))).((((........))))...(((((((((....)))))))))..... ( -39.60, z-score = -1.43, R) >droEre2.scaffold_4784 19867074 91 - 25762168 ACCUCGCCACUUGCGUGCCACCGUG----CCACCCCGACCACACGGCCAGUGGA-GCUGCUCCUGUGGCAGGAGGC----ACCUCUGUGGUGUCCAAGUG .......((((((.(..((((.(((----((...((..(((((.((.(((....-.)))..)))))))..)).)))----))....))))..).)))))) ( -38.30, z-score = -0.88, R) >droAna3.scaffold_13337 6262242 88 - 23293914 ACCUGGGCACU---GAGCCACCGGG----CCACCGGAUAUACCAUACCAUAUACCGUCUCCUCCCUGACUCAGUG-----GCCUGGCUGGUGGCCAAGUG ((.(((.((((---.(((((...((----((((.((((((........)))).))(((........)))...)))-----)))))))))))).))).)). ( -34.90, z-score = -1.66, R) >consensus ACCUCGCCACUUGCGAGCCACCCAG____CCACC__GUCCACCCGGCGGGUGGCUGCCCCUCCUGUGGCAGAAGGG____ACCUCUGUGGUGUCCAAGUG .......((((((...(((((..........................((((....))))..........((.(((......))))))))))...)))))) (-14.18 = -14.30 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:12 2011