| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,125,399 – 20,125,519 |
| Length | 120 |
| Max. P | 0.532960 |

| Location | 20,125,399 – 20,125,519 |
|---|---|
| Length | 120 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.92 |
| Shannon entropy | 0.65410 |
| G+C content | 0.52045 |
| Mean single sequence MFE | -39.66 |
| Consensus MFE | -10.27 |
| Energy contribution | -10.05 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.79 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.532960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
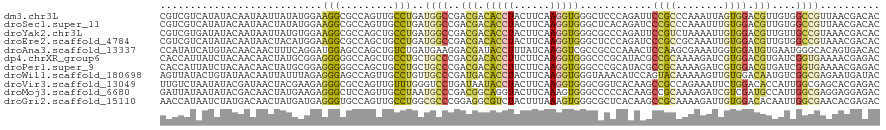
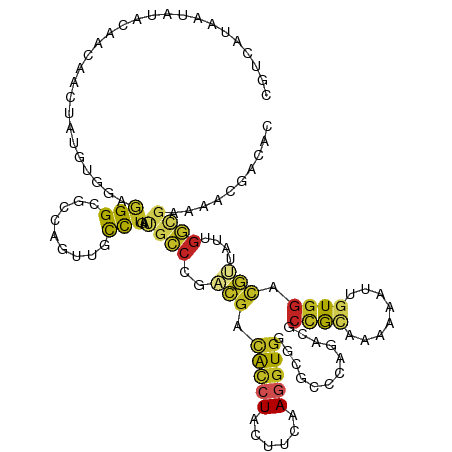
>dm3.chr3L 20125399 120 - 24543557 CGUCGUCAUAUACAAUAAUUAUAUGGAAGGCGCCAGUUGCCUGAUGGCCGACGACACCUACUUCAAGGUGGGCUCCCAGAUUCCGCCCAAAUUAGUGGACGUUGUGGCCGUUAACGACAC .(((((((((((.......))))))..(((((.....)))))(((((((.(((((..(((((......(((((...........)))))....)))))..)))))))))))).))))).. ( -44.00, z-score = -3.16, R) >droSec1.super_11 36138 120 - 2888827 CGUCGUCAUAUACAAUAACUAUAUGGAAGGCGCCAGUUGCCUGAUGGCCGACGACACCUACUUCAAGGUGGGCUCACAGAUCCCGCCCAAAUUUGUGGACGUUGUGGCCGUUAACGACAC .(((((((((((.......))))))..(((((.....)))))(((((((.(((((..((((.....((((((.((...)).)))))).......))))..)))))))))))).))))).. ( -44.50, z-score = -3.10, R) >droYak2.chr3L 3749313 120 + 24197627 CGUCGUGAUAUACAAUAAUUAUGUGGAAGGCGCCAGCUGCCUGAUGGCCGACGACACCUACUUCAAGGUGGGCGCCCAGAUUCCGUCUAAAAUUGUGGACGUUGUUGCCGUAAACGACAC .(((((..(((.(((((((.....((((((((((.((.(((....))).).)..(((((......)))))))))))....))))(((((......))))))))))))..))).))))).. ( -44.40, z-score = -3.13, R) >droEre2.scaffold_4784 19853312 120 - 25762168 CGUCGUCAUAUACAAUAACUACAUGGAAGGCGCCAGCUGCCUGAUGGCCGACGACACCUACUUCAAGGUGGGCUCCCAGAUCCCGCCGCAAAUUGUGGACGUUGUGGCCGUAAACGACAC .(((((.....................(((((.....))))).((((((.(((((..((((.....((((((.((...)).)))))).......))))..)))))))))))..))))).. ( -42.30, z-score = -1.94, R) >droAna3.scaffold_13337 6248095 120 - 23293914 CCAUAUCAUGUACAACAACUUUCAGGAUGGAGCCAGCUGUCUGAUGAAGGACGAUACCUUUAUCAAGGUCGCCGCCCAAACUCCAAGCGAAAUGGUGGAUGUGAAUGGGCACAGUGACAC ..................(((...((((((......))))))((((((((......)))))))))))(((((.(((((.((((((..(.....).)))).))...)))))...))))).. ( -32.50, z-score = -0.38, R) >dp4.chrXR_group6 2000553 120 - 13314419 CACCAUUAUCUACAACAACUAUGCGGAGGGGGCCAGCUGCCUGCUGCCCGACGACACCUUCUUCAAGGUGGGCCCGCAUACGCCGCAAAAGAUCGUGGACGUGAUCGGUGAAAACGAGAC ((((.................(((((...((((.(((.....))))))).....((((((....))))))...)))))((((((((........)))).))))...)))).......... ( -42.20, z-score = -1.80, R) >droPer1.super_9 293482 120 - 3637205 CACCAUUAUCUACAACAACUAUGCGGAGGGGGCCAGCUGCCUGCUGCCCGACGACACCUUCUUCAAGGUGGGCCCGCAUACGCCGCAAAAGAUCGUGGACGUGAUCGGUGAAAACGAGAC ((((.................(((((...((((.(((.....))))))).....((((((....))))))...)))))((((((((........)))).))))...)))).......... ( -42.20, z-score = -1.80, R) >droWil1.scaffold_180698 8177839 120 + 11422946 AGUUAUACUGUAUAACAAUUAUUUAGAGGGAGCCAGUUGCCUGUUGCCCGAUGACACCUACUUCAAGGUGGGUAAACAUCCAGUACAAAAAGUUGUGGACAAUGUCGGCGAGAAUGAUAC .((((((...)))))).(((((((..(((.((....)).))).(((((.(((...(((((((....))))))).....((((..((.....))..))))....))))))))))))))).. ( -29.60, z-score = -0.54, R) >droVir3.scaffold_13049 6692749 120 + 25233164 UUGUCUAAUAUACGAUAACUACGAAGAGGGCGCCAGUUGUUUGGGUCCUGAUAAUACCUACUUCAAGGUGGGCGGUCACAAGCCGCCAGAAAUUCUGGACACCAUUGGCGAGCACGAGAC ..((((......((.......))....(..((((((.....(((((((.((...(((((......)))))((((((.....))))))......)).)))).)))))))))..)...)))) ( -36.80, z-score = -1.49, R) >droMoj3.scaffold_6680 5423156 120 + 24764193 GAUUAUAAUAUACGACAACUAUGAAGAGGGCUCCAGUUGCCUAAUGCCCGACGGCAGGUACUUCAAAGUGGGCCCCCACAAGCCGCAAAAGAUCGUCGAUGCCAUUGGCGAGGAGGAGAC ............((((.....(((((.((((.......))))..((((....))))....)))))..((((....))))...............)))).((((...)))).......... ( -31.00, z-score = 0.20, R) >droGri2.scaffold_15110 9113507 120 - 24565398 AACCAUAAUCUAUGACAACUAUGAUGAGGGUGCCAGUUGCCUGGCGCCCGGAGGCGUCUACUUUAAAGUGGGCGCUCACAAGCCGCAAAAGAUUGUGGACACAAUUGGCGAACACGAGAC ..(((((((((................(((((((((....)))))))))(..((((((((((....))))))))))..)..........)))))))))...................... ( -46.80, z-score = -4.09, R) >consensus CGUCAUAAUAUACAACAACUAUGUGGAGGGCGCCAGUUGCCUGAUGCCCGACGACACCUACUUCAAGGUGGGCGCCCAGACGCCGCAAAAAAUUGUGGACGUUAUUGGCGAAAACGACAC ...........................(((.........)))..((((..(((.(((((......)))))............((((........)))).)))....)))).......... (-10.27 = -10.05 + -0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:09 2011