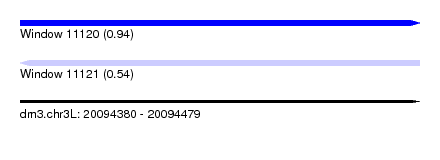
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,094,380 – 20,094,479 |
| Length | 99 |
| Max. P | 0.940521 |

| Location | 20,094,380 – 20,094,479 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 69.81 |
| Shannon entropy | 0.59958 |
| G+C content | 0.69372 |
| Mean single sequence MFE | -42.44 |
| Consensus MFE | -26.40 |
| Energy contribution | -27.20 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
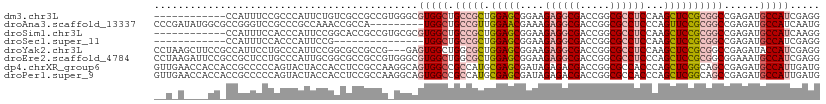
>dm3.chr3L 20094380 99 + 24543557 ------------CCAUUUCCGCCCAUUCUGUCGCCGCCGUGGGCGUGGCUGCCGCUGGAGCGGAAGAGGCGACCGGCGCCUCCAAGCUCCGCGGCCGAGAUGCCAUCGAGG ------------((.....(((((((..((....))..)))))))((((.(((((.(((((....((((((.....))))))...))))))))))......))))....)) ( -46.80, z-score = -0.91, R) >droAna3.scaffold_13337 18254279 102 - 23293914 CCCGAUAUGGCGCCGGGUCCGCCCGCCAAACCGCCA---------UGGCUGCCGUUGGAACGAAAGAGGCGACCGGCGCCUCCCAGUUCCGCGGCCGAGAUGCCAUCAAUG ...(((((((((.((((....))))......)))))---------)(((.(((((.(((((....((((((.....))))))...))))))))))......)))))).... ( -45.10, z-score = -2.06, R) >droSim1.chr3L 19389172 99 + 22553184 ------------CCAUUUCCACCCAUUCCGGCACCGCCGUGCGCGUGGCUGCCGCUGGAGCGGAAGAGGCGACCGGCGCCUCCAAGCUCCGCGGCCGAGAUGCCAUCAAGG ------------.((((((..........(.((((((...))).))).).(((((.(((((....((((((.....))))))...)))))))))).))))))......... ( -43.00, z-score = -0.97, R) >droSec1.super_11 10390 84 + 2888827 ------------CCAUUUCCACCCAUUCCG---------------UGGCUGCCGCUGGAGCGGAAGAGGCGACCGGCGCCUCCAAGCUCCGCGGCCGAGAUGCCAUCGAGG ------------.............(((.(---------------((((.(((((.(((((....((((((.....))))))...))))))))))......))))).))). ( -38.70, z-score = -2.15, R) >droYak2.chr3L 3729271 108 - 24197627 CCUAAGCUUCCGCCAUUCCUGCCCAUUCCGGCGCCGCCG---GAGUGGCUGGCGCUGGAGCGGAAGAGGCGACCGGCGCCUCCAAGCUCCGCGGCCGAGAUACCAUCGAGG (((........(((...((.(((.((((((((...))))---))))))).)).((.(((((....((((((.....))))))...))))))))))(((.......)))))) ( -50.40, z-score = -1.25, R) >droEre2.scaffold_4784 19832131 111 + 25762168 CCUAAGAUUCCGCCGCUCCUGCCCAUUGCGGCGCCGCCGUGGGCGUGGCUGGCGCUGGAGCGGAAGAGGCGACCGGCGCCUCCCAGCUCCGCGGCGGAAAUGCCAUCGAGG .......(((((((((....((((((.(((....))).))))))(..((((((((....)))...((((((.....)))))))))))..))))))))))............ ( -60.50, z-score = -1.95, R) >dp4.chrXR_group6 4677684 111 + 13314419 GUUGAACCACCACCGCCCCCAGUACUACCACCUCCGCCAAGGCAGUGGCCGCCAUGCGAGCGAUAGAGACGACCGGCGCCACCCAGCUCGGCAGCCGAGAUGCCAUUGAUG .....................................((((((((((((.(((...((..(....)...))...))))))))....(((((...))))).)))).)))... ( -27.50, z-score = 0.49, R) >droPer1.super_9 2978343 111 + 3637205 GUUGAACCACCACCGCCCCCAGUACUACCACCUCCGCCAAGGCAGUGGCCGCCAUGCGAGCGAUAGAGACGACCGGCGCCACCCAGCUCGGCAGCCGAGAUGCCAUUGAUG .....................................((((((((((((.(((...((..(....)...))...))))))))....(((((...))))).)))).)))... ( -27.50, z-score = 0.49, R) >consensus __U_A____CC_CCAUUCCCACCCAUUCCGCCGCCGCCG_GGGAGUGGCUGCCGCUGGAGCGGAAGAGGCGACCGGCGCCUCCAAGCUCCGCGGCCGAGAUGCCAUCGAGG ............................................(((((.(((((.(((((....((((((.....))))))...))))))))))......)))))..... (-26.40 = -27.20 + 0.80)

| Location | 20,094,380 – 20,094,479 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.81 |
| Shannon entropy | 0.59958 |
| G+C content | 0.69372 |
| Mean single sequence MFE | -47.81 |
| Consensus MFE | -26.40 |
| Energy contribution | -25.34 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.61 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.544476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
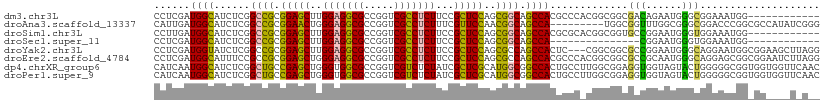
>dm3.chr3L 20094380 99 - 24543557 CCUCGAUGGCAUCUCGGCCGCGGAGCUUGGAGGCGCCGGUCGCCUCUUCCGCUCCAGCGGCAGCCACGCCCACGGCGGCGACAGAAUGGGCGGAAAUGG------------ ......((((......((((((((((..(((((((.....)))))))...))))).))))).))))((((((....(....)....)))))).......------------ ( -47.70, z-score = -0.94, R) >droAna3.scaffold_13337 18254279 102 + 23293914 CAUUGAUGGCAUCUCGGCCGCGGAACUGGGAGGCGCCGGUCGCCUCUUUCGUUCCAACGGCAGCCA---------UGGCGGUUUGGCGGGCGGACCCGGCGCCAUAUCGGG .....(((((......((((.(((((.((((((((.....))))))))..)))))..)))).))))---------)..((((.((((.(.((....)).))))).)))).. ( -45.60, z-score = -0.33, R) >droSim1.chr3L 19389172 99 - 22553184 CCUUGAUGGCAUCUCGGCCGCGGAGCUUGGAGGCGCCGGUCGCCUCUUCCGCUCCAGCGGCAGCCACGCGCACGGCGGUGCCGGAAUGGGUGGAAAUGG------------ (((...(((((((...((((((((((..(((((((.....)))))))...))))).))))).(((........))))))))))....))).........------------ ( -50.50, z-score = -1.44, R) >droSec1.super_11 10390 84 - 2888827 CCUCGAUGGCAUCUCGGCCGCGGAGCUUGGAGGCGCCGGUCGCCUCUUCCGCUCCAGCGGCAGCCA---------------CGGAAUGGGUGGAAAUGG------------ (((((.((((......((((((((((..(((((((.....)))))))...))))).))))).))))---------------)))...))..........------------ ( -39.00, z-score = -0.86, R) >droYak2.chr3L 3729271 108 + 24197627 CCUCGAUGGUAUCUCGGCCGCGGAGCUUGGAGGCGCCGGUCGCCUCUUCCGCUCCAGCGCCAGCCACUC---CGGCGGCGCCGGAAUGGGCAGGAAUGGCGGAAGCUUAGG (((............(((.(((((((..(((((((.....)))))))...))))).))))).(((..((---((((...))))))...))))))...(((....))).... ( -51.30, z-score = -0.26, R) >droEre2.scaffold_4784 19832131 111 - 25762168 CCUCGAUGGCAUUUCCGCCGCGGAGCUGGGAGGCGCCGGUCGCCUCUUCCGCUCCAGCGCCAGCCACGCCCACGGCGGCGCCGCAAUGGGCAGGAGCGGCGGAAUCUUAGG (((....((...((((((((((..(((((((((((.....))))))...(((....))))))))..)(((((..(((....)))..)))))....))))))))).)).))) ( -58.80, z-score = -1.58, R) >dp4.chrXR_group6 4677684 111 - 13314419 CAUCAAUGGCAUCUCGGCUGCCGAGCUGGGUGGCGCCGGUCGUCUCUAUCGCUCGCAUGGCGGCCACUGCCUUGGCGGAGGUGGUAGUACUGGGGGCGGUGGUGGUUCAAC .((((..(((..(.((((.((((.......)))))))))..))).(((((((((.((..((.((((((.((.....)).)))))).))..)).)))))))))))))..... ( -44.80, z-score = 0.26, R) >droPer1.super_9 2978343 111 - 3637205 CAUCAAUGGCAUCUCGGCUGCCGAGCUGGGUGGCGCCGGUCGUCUCUAUCGCUCGCAUGGCGGCCACUGCCUUGGCGGAGGUGGUAGUACUGGGGGCGGUGGUGGUUCAAC .((((..(((..(.((((.((((.......)))))))))..))).(((((((((.((..((.((((((.((.....)).)))))).))..)).)))))))))))))..... ( -44.80, z-score = 0.26, R) >consensus CCUCGAUGGCAUCUCGGCCGCGGAGCUGGGAGGCGCCGGUCGCCUCUUCCGCUCCAGCGGCAGCCACGCCC_CGGCGGCGCCGGAAUGGGCGGAAACGG_GG____U_A__ ......((((......((((.(((((..(((((((.....)))))))...)))))..)))).)))).............(((......))).................... (-26.40 = -25.34 + -1.06)
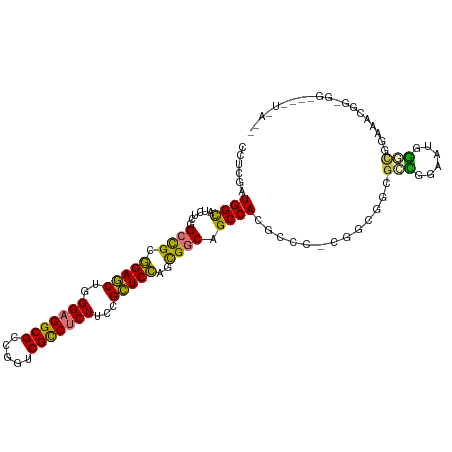
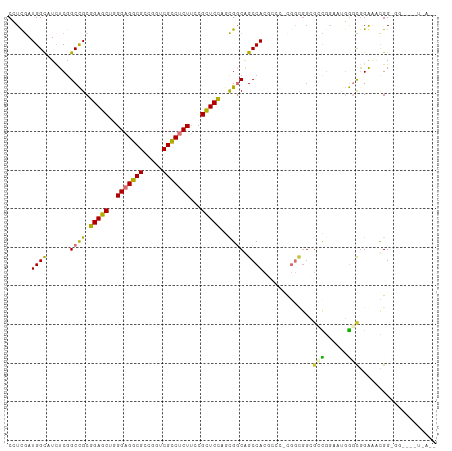
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:39:05 2011