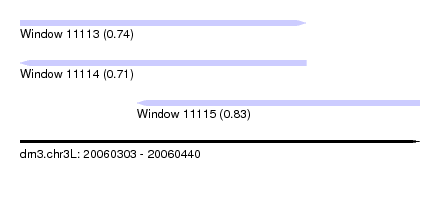
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,060,303 – 20,060,440 |
| Length | 137 |
| Max. P | 0.830179 |

| Location | 20,060,303 – 20,060,401 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 69.26 |
| Shannon entropy | 0.59620 |
| G+C content | 0.57419 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -13.43 |
| Energy contribution | -15.09 |
| Covariance contribution | 1.66 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.744494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
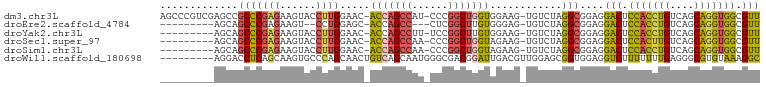
>dm3.chr3L 20060303 98 + 24543557 GCCAAGCGAACUUUUCCUUGACGCCCUUUAAGUCCCUGGCAACGCCACCUGCUGACAGGUGGAGUCCUCC--GCCUAGACACUUCCACCAGCCGGGAUGG----------- ..((((.((.....))))))...........(((((.(((....(((((((....))))))).(((....--.....)))..........))))))))..----------- ( -29.60, z-score = -0.90, R) >droAna3.scaffold_13337 18222811 110 - 23293914 GCCGGGCCAACUUUUCCUUGACGCC-UUUAAGUCCUUGGCAACACCACCUGCUGACAGGAGGAGUUCUCCCAAAUAAUAAAAAAAAAAAAGCCAAGAACCACCCGACCUCC ..((((.............(((...-.....)))((((((.......((((....)))).(((....)))....................)))))).....))))...... ( -26.10, z-score = -1.94, R) >droEre2.scaffold_4784 19800023 95 + 25762168 ACCAAGCGAACU-UUCCUUGACGCCCUCUAAGUCCCUGGCAACGCCACCUGCUGACAGGUGGAGUCCUCC--GCCUAGACACUCCCACAAGCCGAGGG------------- ..((((.((...-.))))))...(((((...(((...(((.((.(((((((....))))))).)).....--)))..))).((......))..)))))------------- ( -28.90, z-score = -1.25, R) >droYak2.chr3L 3694052 98 - 24197627 GCCAAGCGAACUUUUCCUUGACGCCCUUUAAGUCCCUGGCAACGCCACCUGCUGACAGGUGGAGUCCUCC--GCCUAGACACUUCCACAAGCCGGAAAGG----------- ..........(((((((..(((.........)))...(((....(((((((....))))))).(((....--.....)))..........))))))))))----------- ( -27.50, z-score = -0.76, R) >droSec1.super_97 53141 98 + 93343 GCCAGGCGAACUGUUCCUUGACGCCCUUUAAGUCCCUGGCAACGCCACCUGCUGACAAGUGGAGUCCUCC--GCCUAGACACUUCUACCAGCCGGGUUGG----------- ((((((.((.......(((((......)))))))))))))....((((((((((....(((((....)))--)).((((....)))).))).)))).)))----------- ( -30.91, z-score = -0.76, R) >droSim1.chr3L 19353331 98 + 22553184 GCCGAGCGAACUUUUCCUUGACGCCCUUUAAGUCCCUGGCAACGCCACCUGCUGACAGGUGGAGUCCUCC--GCCUAGACACUUCUACCAGCCGGGUUGG----------- ((((((.((.....)))))(((.........)))...)))....((((((((((..(((((((....)))--))))(((....)))..))).)))).)))----------- ( -29.20, z-score = -0.42, R) >droWil1.scaffold_180698 1214438 83 + 11422946 ----------------CAGCAAACUUUUUGAGGCCCUU-CGCCUUUACACGCCCUCAAAAAAAGACCUCCACCGCUCCA-ACGUCAAUCCGUCGCCCAUUG---------- ----------------..((....(((((((((.....-.((........)))))))))))..(((......((.....-.)).......)))))......---------- ( -11.32, z-score = -1.07, R) >consensus GCCAAGCGAACUUUUCCUUGACGCCCUUUAAGUCCCUGGCAACGCCACCUGCUGACAGGUGGAGUCCUCC__GCCUAGACACUUCCACCAGCCGGGAAGG___________ ..................................((((((....(((((((....))))))).(((...........)))..........))))))............... (-13.43 = -15.09 + 1.66)

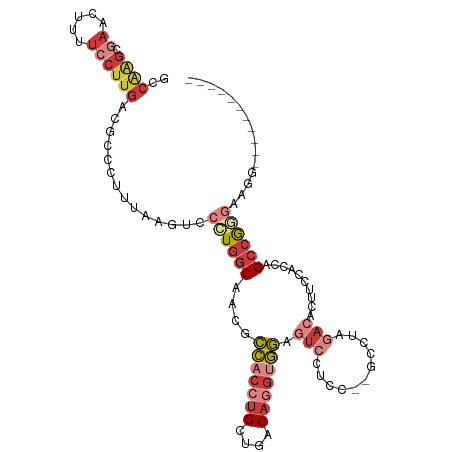
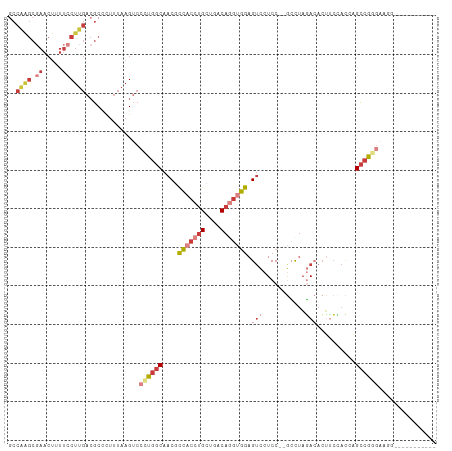
| Location | 20,060,303 – 20,060,401 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.26 |
| Shannon entropy | 0.59620 |
| G+C content | 0.57419 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -20.38 |
| Energy contribution | -20.25 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.68 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20060303 98 - 24543557 -----------CCAUCCCGGCUGGUGGAAGUGUCUAGGC--GGAGGACUCCACCUGUCAGCAGGUGGCGUUGCCAGGGACUUAAAGGGCGUCAAGGAAAAGUUCGCUUGGC -----------(((((......)))))((((.(((.(((--....(((.(((((((....))))))).))))))))).)))).......((((((((.....)).)))))) ( -36.70, z-score = -0.58, R) >droAna3.scaffold_13337 18222811 110 + 23293914 GGAGGUCGGGUGGUUCUUGGCUUUUUUUUUUUUAUUAUUUGGGAGAACUCCUCCUGUCAGCAGGUGGUGUUGCCAAGGACUUAAA-GGCGUCAAGGAAAAGUUGGCCCGGC ....(((((((((((((((((.......((((((.....))))))(((.((.((((....)))).)).)))))))))))))....-(((...........))).))))))) ( -37.50, z-score = -1.47, R) >droEre2.scaffold_4784 19800023 95 - 25762168 -------------CCCUCGGCUUGUGGGAGUGUCUAGGC--GGAGGACUCCACCUGUCAGCAGGUGGCGUUGCCAGGGACUUAGAGGGCGUCAAGGAA-AGUUCGCUUGGU -------------(((((.((((....))))((((.(((--....(((.(((((((....))))))).))))))..))))...)))))..(((((((.-...)).))))). ( -36.80, z-score = -0.47, R) >droYak2.chr3L 3694052 98 + 24197627 -----------CCUUUCCGGCUUGUGGAAGUGUCUAGGC--GGAGGACUCCACCUGUCAGCAGGUGGCGUUGCCAGGGACUUAAAGGGCGUCAAGGAAAAGUUCGCUUGGC -----------........((((....((((.(((.(((--....(((.(((((((....))))))).))))))))).))))...))))((((((((.....)).)))))) ( -37.10, z-score = -0.95, R) >droSec1.super_97 53141 98 - 93343 -----------CCAACCCGGCUGGUAGAAGUGUCUAGGC--GGAGGACUCCACUUGUCAGCAGGUGGCGUUGCCAGGGACUUAAAGGGCGUCAAGGAACAGUUCGCCUGGC -----------.....(((.((((.........)))).)--))..(((.(((((((....))))))).)))(((((((((.........)))..(((....))).)))))) ( -34.40, z-score = 0.04, R) >droSim1.chr3L 19353331 98 - 22553184 -----------CCAACCCGGCUGGUAGAAGUGUCUAGGC--GGAGGACUCCACCUGUCAGCAGGUGGCGUUGCCAGGGACUUAAAGGGCGUCAAGGAAAAGUUCGCUCGGC -----------((..((((.((((.........)))).)--((..(((.(((((((....))))))).))).)).))).......(((((.............))))))). ( -35.92, z-score = -0.57, R) >droWil1.scaffold_180698 1214438 83 - 11422946 ----------CAAUGGGCGACGGAUUGACGU-UGGAGCGGUGGAGGUCUUUUUUUGAGGGCGUGUAAAGGCG-AAGGGCCUCAAAAAGUUUGCUG---------------- ----------.....(((((((......)))-).((((....((((((((((.......((.(....).)))-))))))))).....))))))).---------------- ( -22.71, z-score = -0.58, R) >consensus ___________CCAUCCCGGCUGGUGGAAGUGUCUAGGC__GGAGGACUCCACCUGUCAGCAGGUGGCGUUGCCAGGGACUUAAAGGGCGUCAAGGAAAAGUUCGCUUGGC ...............((((((...((((....)))).........(((.(((((((....))))))).)))))).))).......(((((.............)))))... (-20.38 = -20.25 + -0.13)
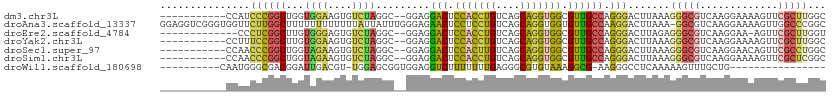


| Location | 20,060,343 – 20,060,440 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 75.58 |
| Shannon entropy | 0.43831 |
| G+C content | 0.60443 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -25.47 |
| Energy contribution | -24.90 |
| Covariance contribution | -0.57 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20060343 97 - 24543557 AGCCCGUCGAGCCGCCCGAGAAGUACCUUGGAAC-ACCAGCCAU-CCCGGCUGGUGGAAG-UGUCUAGGCGGAGGACUCCACCUGUCAGCAGGUGGCGUU ...(((((.((.((((((((......)))))..(-(((((((..-...))))))))...)-)).)).)))))..(((.(((((((....))))))).))) ( -48.90, z-score = -3.70, R) >droEre2.scaffold_4784 19800062 84 - 25762168 ---------AGCAGCCCGAGAAGU--CCUGGAGC-ACCAGCC---CUCGGCUUGUGGGAG-UGUCUAGGCGGAGGACUCCACCUGUCAGCAGGUGGCGUU ---------.((.((((.(.((((--(..((.((-....)))---)..))))).).)).)-)......))....(((.(((((((....))))))).))) ( -32.40, z-score = 0.25, R) >droYak2.chr3L 3694092 88 + 24197627 ---------AGCAGCCCGAGAAGUACCUUGGAAC-ACCAGCCUU-UCCGGCUUGUGGAAG-UGUCUAGGCGGAGGACUCCACCUGUCAGCAGGUGGCGUU ---------.((...(((((......)))))..(-((.((((..-...)))).)))....-.......))....(((.(((((((....))))))).))) ( -32.70, z-score = -0.67, R) >droSec1.super_97 53181 88 - 93343 ---------AGCAGCCCGAGAAGUACCUUGGAAC-ACCAGCCAA-CCCGGCUGGUAGAAG-UGUCUAGGCGGAGGACUCCACUUGUCAGCAGGUGGCGUU ---------.((...(((((......)))))...-(((((((..-...))))))).....-.......))....(((.(((((((....))))))).))) ( -33.20, z-score = -1.22, R) >droSim1.chr3L 19353371 88 - 22553184 ---------AGCAGCCCGAGAAGUACCUUGGAAC-ACCAGCCAA-CCCGGCUGGUAGAAG-UGUCUAGGCGGAGGACUCCACCUGUCAGCAGGUGGCGUU ---------.((...(((((......)))))...-(((((((..-...))))))).....-.......))....(((.(((((((....))))))).))) ( -35.90, z-score = -1.87, R) >droWil1.scaffold_180698 1214461 91 - 11422946 ---------AGGACCUCAGCAAGUGCCCAGCAACUGUCAGCAAUGGGCGACGGAUUGACGUUGGAGCGGUGGAGGUCUUUUUUUGAGGGCGUGUAAAGGC ---------((((((((....(.((((((((..(((((.((.....)))))))......))))).))).).)))))))).........((.(....).)) ( -27.90, z-score = -0.06, R) >consensus _________AGCAGCCCGAGAAGUACCUUGGAAC_ACCAGCCAU_CCCGGCUGGUGGAAG_UGUCUAGGCGGAGGACUCCACCUGUCAGCAGGUGGCGUU ...............(((........((((((...(((((((......)))))))........)))))))))..(((.(((((((....))))))).))) (-25.47 = -24.90 + -0.57)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:59 2011