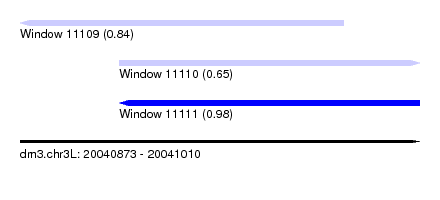
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,040,873 – 20,041,010 |
| Length | 137 |
| Max. P | 0.983635 |

| Location | 20,040,873 – 20,040,984 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 64.70 |
| Shannon entropy | 0.69775 |
| G+C content | 0.49614 |
| Mean single sequence MFE | -33.79 |
| Consensus MFE | -13.07 |
| Energy contribution | -14.79 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.841098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
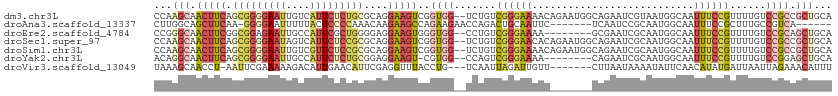
>dm3.chr3L 20040873 111 - 24543557 CCAAGCAACUUCAGCGGGGAAUUGUCAUUCUCUGCGCAGGAAGUCGGUGG--UCUGUCGGGAAAACAGAAUGGCAGAAUCGUAAUGGCAAUUUCCGUUUUGUCCGCCGCUGCA ....((((((((.(((((((((....)))))))))....)))))((((((--.(...((((((..((..((((.....))))..))....))))))....).)))))).))). ( -38.00, z-score = -1.47, R) >droAna3.scaffold_13337 18202912 99 + 23293914 CUUGGCAGCUUCAA-GGGGAAUUUUUACUCCCCAAACAAGAAGCCAGAAGAACCAGACUGCAAUUC-------UCAAUCCGCAAUGGCAAUUUCCGCUUUGCCGUCA------ .(((((..(((...-(((((........)))))....)))..)))))........((((((.....-------.......)))..(((((........)))))))).------ ( -27.20, z-score = -1.61, R) >droEre2.scaffold_4784 19780681 103 - 25762168 CCGGGCAACUUCGGCGGAGAAUUGCCAUUCGCUGGGGAGGAAGUGGGUGG--CCUGUCGGGAAAA--------GCGAAUCGCAAUGGCAAUUUCCGUUUUGUCCGCAGCUGCA ..((((((....((((((((.((((((((..((.((.(((..(....)..--))).)).))....--------(((...)))))))))))))))))))))))))((....)). ( -37.50, z-score = -0.19, R) >droSec1.super_97 33643 111 - 93343 CCAAGCAACUUCAGCGGGGAAUAGUCAUUCUCCGCGCAGGAAGUCGGUGG--UCUGUCGGGAACACAGAAUGGCAGAAUCGCAAUGGCAAUUUCCGUUUUGUCCGCCGCUGCA ....((((((((.(((((((((....)))))))))....)))))((((((--......(....)(((((((((..((((.((....)).))))))))))))))))))).))). ( -42.40, z-score = -2.42, R) >droSim1.chr3L 19333531 111 - 22553184 CCAAGCAACUUCAGCGGGGAAUUGUCGUUCUCCGCGCAGGAAGUCGGUGG--UCUGUCGGGAAAACAGAAUGGCAGAAUCGCAAUGGCAAUUUCCGUUUUGUCCGCCGCUGCA ....((((((((.(((((((((....)))))))))....)))))((((((--(((....)))..(((((((((..((((.((....)).))))))))))))))))))).))). ( -41.90, z-score = -1.88, R) >droYak2.chr3L 3670087 102 + 24197627 ACAGGCAACUUCAGCGGGGAAUUGCCAUUCUCUGCGGAGGAAGU-CGUGG--CCAGUCGGGAAAA--------CAGAAUCGCAAUGGCAAUUUCCGUUUUGUCCGGAGCUGCA ...(((..((((.(((((((((....))))))))).))))..))-)(..(--(...(((((((((--------(.(((..((....))...))).))))).))))).))..). ( -37.90, z-score = -1.79, R) >droVir3.scaffold_13049 18219957 102 - 25233164 UAAAGCAACCU-AAUUCGAAAAAGACAUUGAACAUUCGAGGUUUACCUG---UCAAUUAGAUUGUU-------CUUAAUAAAAUAUUCAACAUAUGAUUAAUUAGAAACAUUU ...(((((.((-((((.((...((((.((((....)))).)))).....---)))))))).)))))-------.(((((...((((.....)))).)))))............ ( -11.60, z-score = -0.07, R) >consensus CCAAGCAACUUCAGCGGGGAAUUGUCAUUCUCCGCGCAGGAAGUCGGUGG__UCUGUCGGGAAAAC_______CAGAAUCGCAAUGGCAAUUUCCGUUUUGUCCGCAGCUGCA ...(((.(((((.(((((((((....)))))))))....)))))..((((.......((((((...........................))))))......)))).)))... (-13.07 = -14.79 + 1.72)
| Location | 20,040,907 – 20,041,010 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 65.07 |
| Shannon entropy | 0.67626 |
| G+C content | 0.44089 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -5.88 |
| Energy contribution | -5.54 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.71 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.652944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20040907 103 + 24543557 AUUCUGCCAUUCUGUUUUCCCGACAGA--CCACCGACUUCCUGCGCAGAGAAUGACAAUUCCCCGCUGAAGUUGCUUGGCUAAAAUGCA-UUUAUGACAUUUUUCU .....((((.((((((.....))))))--....(((((((..(((..(.((((....))))).))).)))))))..)))).((((((((-....)).))))))... ( -25.60, z-score = -3.11, R) >droAna3.scaffold_13337 18202940 97 - 23293914 -------GAUUGAGAAUUGCAGUCUGGUUCUUCUGGCUUCUUGUUUGGGGAGUAAAAAUUCCCC-UUGAAGCUGCCAAGCCCGAGAACA-UUUAUGACAUUUUUCU -------....(((((.((((.....((((((..(((((...(((((((((((....)))))))-...))))....))))).)))))).-....)).)).))))). ( -30.40, z-score = -2.42, R) >droEre2.scaffold_4784 19780715 95 + 25762168 --------AUUCGCUUUUCCCGACAGG--CCACCCACUUCCUCCCCAGCGAAUGGCAAUUCUCCGCCGAAGUUGCCCGGCUAAAAUGCA-UUUAUGACAUUUUUCU --------(((((((......((.((.--.......))))......)))))))(((((((.((....))))))))).....((((((((-....)).))))))... ( -16.90, z-score = 0.04, R) >droSec1.super_97 33677 103 + 93343 AUUCUGCCAUUCUGUGUUCCCGACAGA--CCACCGACUUCCUGCGCGGAGAAUGACUAUUCCCCGCUGAAGUUGCUUGGCUAAAAUGCA-UUUAUGACAUUUUUCU .....((((.(((((.......)))))--....(((((((..(((.((.((((....))))))))).)))))))..)))).((((((((-....)).))))))... ( -28.40, z-score = -3.50, R) >droSim1.chr3L 19333565 103 + 22553184 AUUCUGCCAUUCUGUUUUCCCGACAGA--CCACCGACUUCCUGCGCGGAGAACGACAAUUCCCCGCUGAAGUUGCUUGGCUAAAAUGCA-UUUAUGACAUUUUUCU .....((((.((((((.....))))))--....(((((((..(((.((.(((......)))))))).)))))))..)))).((((((((-....)).))))))... ( -28.40, z-score = -3.74, R) >droYak2.chr3L 3670121 94 - 24197627 --------AUUCUGUUUUCCCGACUGG--CCACG-ACUUCCUCCGCAGAGAAUGGCAAUUCCCCGCUGAAGUUGCCUGUCUAAAAUGCA-UUUAUGACAUUUUUCU --------.............(((.((--(...(-(((((....((.(.((((....)))).).)).))))))))).))).((((((((-....)).))))))... ( -19.30, z-score = -1.06, R) >droVir3.scaffold_13049 18219997 90 + 25233164 -------------AACAAUCUAAUUGA---CAGGUAAACCUCGAAUGUUCAAUGUCUUUUUCGAAUUAGGUUGCUUUAUUUCAAAGGCAAUUUGUGAUAACACCUU -------------..............---.((((.....(((((..............)))))..(((((((((((......))))))))))).......)))). ( -15.04, z-score = -0.53, R) >consensus ________AUUCUGUUUUCCCGACAGA__CCACCGACUUCCUGCGCGGAGAAUGACAAUUCCCCGCUGAAGUUGCUUGGCUAAAAUGCA_UUUAUGACAUUUUUCU .................................(((((((....((.(.((((....)))).).)).)))))))................................ ( -5.88 = -5.54 + -0.34)


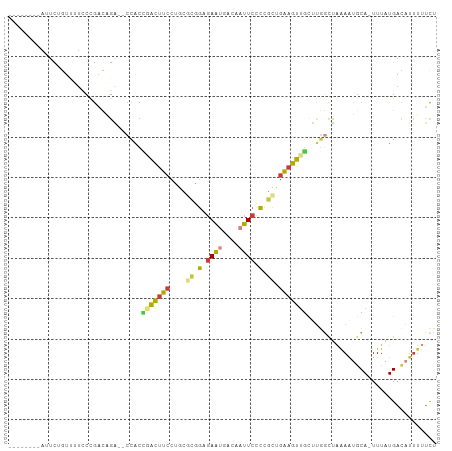
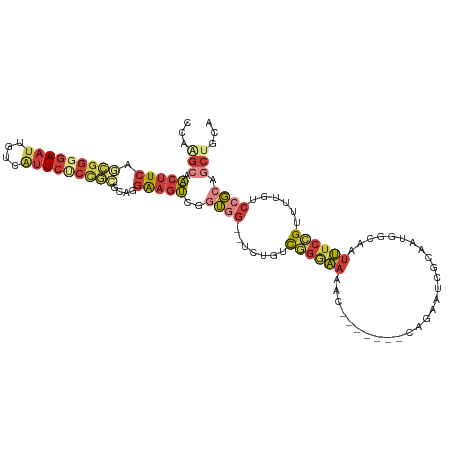
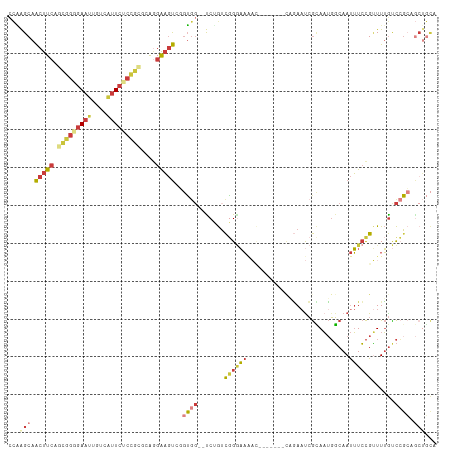
| Location | 20,040,907 – 20,041,010 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 65.07 |
| Shannon entropy | 0.67626 |
| G+C content | 0.44089 |
| Mean single sequence MFE | -26.69 |
| Consensus MFE | -12.73 |
| Energy contribution | -12.33 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.76 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20040907 103 - 24543557 AGAAAAAUGUCAUAAA-UGCAUUUUAGCCAAGCAACUUCAGCGGGGAAUUGUCAUUCUCUGCGCAGGAAGUCGGUGG--UCUGUCGGGAAAACAGAAUGGCAGAAU ...(((((((......-.))))))).((((.((.(((((.(((((((((....)))))))))....)))))..))..--(((((.......))))).))))..... ( -29.80, z-score = -2.02, R) >droAna3.scaffold_13337 18202940 97 + 23293914 AGAAAAAUGUCAUAAA-UGUUCUCGGGCUUGGCAGCUUCAA-GGGGAAUUUUUACUCCCCAAACAAGAAGCCAGAAGAACCAGACUGCAAUUCUCAAUC------- .((.((.(((......-.((((((.(((((((.....))..-(((((........))))).......))))).).)))))......))).)).))....------- ( -20.82, z-score = -0.24, R) >droEre2.scaffold_4784 19780715 95 - 25762168 AGAAAAAUGUCAUAAA-UGCAUUUUAGCCGGGCAACUUCGGCGGAGAAUUGCCAUUCGCUGGGGAGGAAGUGGGUGG--CCUGUCGGGAAAAGCGAAU-------- ................-(((.((((.((((((....))))))...((...((((((((((........)))))))))--)...))..)))).)))...-------- ( -27.70, z-score = -0.58, R) >droSec1.super_97 33677 103 - 93343 AGAAAAAUGUCAUAAA-UGCAUUUUAGCCAAGCAACUUCAGCGGGGAAUAGUCAUUCUCCGCGCAGGAAGUCGGUGG--UCUGUCGGGAACACAGAAUGGCAGAAU ...(((((((......-.))))))).((((.((.(((((.(((((((((....)))))))))....)))))..))..--(((((.(....)))))).))))..... ( -34.50, z-score = -3.56, R) >droSim1.chr3L 19333565 103 - 22553184 AGAAAAAUGUCAUAAA-UGCAUUUUAGCCAAGCAACUUCAGCGGGGAAUUGUCGUUCUCCGCGCAGGAAGUCGGUGG--UCUGUCGGGAAAACAGAAUGGCAGAAU ...(((((((......-.))))))).((((.((.(((((.(((((((((....)))))))))....)))))..))..--(((((.......))))).))))..... ( -32.30, z-score = -2.48, R) >droYak2.chr3L 3670121 94 + 24197627 AGAAAAAUGUCAUAAA-UGCAUUUUAGACAGGCAACUUCAGCGGGGAAUUGCCAUUCUCUGCGGAGGAAGU-CGUGG--CCAGUCGGGAAAACAGAAU-------- ...(((((((......-.))))))).(((.(((.(((((.(((((((((....)))))))))....)))))-....)--)).))).............-------- ( -28.10, z-score = -2.30, R) >droVir3.scaffold_13049 18219997 90 - 25233164 AAGGUGUUAUCACAAAUUGCCUUUGAAAUAAAGCAACCUAAUUCGAAAAAGACAUUGAACAUUCGAGGUUUACCUG---UCAAUUAGAUUGUU------------- ((((((...........))))))........(((((.((((((.((...((((.((((....)))).)))).....---)))))))).)))))------------- ( -13.60, z-score = -0.07, R) >consensus AGAAAAAUGUCAUAAA_UGCAUUUUAGCCAAGCAACUUCAGCGGGGAAUUGUCAUUCUCCGCGCAGGAAGUCGGUGG__UCUGUCGGGAAAACAGAAU________ ...........................(((....(((((.(((((((((....)))))))))....)))))...)))............................. (-12.73 = -12.33 + -0.40)


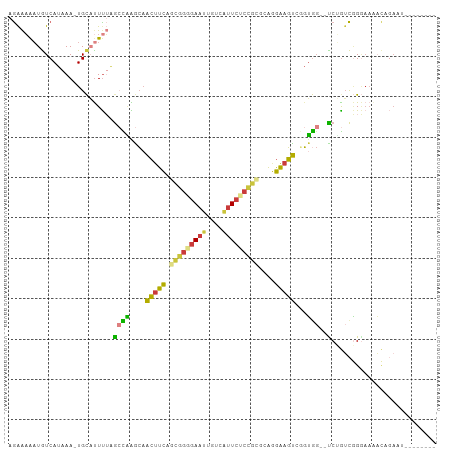
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:56 2011