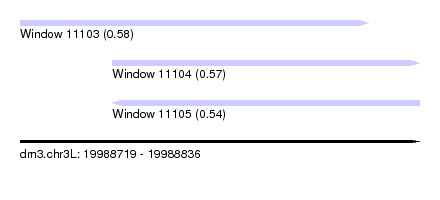
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,988,719 – 19,988,836 |
| Length | 117 |
| Max. P | 0.580039 |

| Location | 19,988,719 – 19,988,821 |
|---|---|
| Length | 102 |
| Sequences | 14 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.61 |
| Shannon entropy | 0.52201 |
| G+C content | 0.42402 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -13.72 |
| Energy contribution | -13.33 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.79 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19988719 102 + 24543557 ---------------UCUUCUCAGGCAAUUUAUGACAUGCUAGGAGCGUGCUCAUCUAAUCGUCCAGCUGAUUCAGCAGAAGAAAGAGCA-AAAAAUAUUUUUGCAAAAAUGGACGAA ---------------................(((((((((.....))))).))))....(((((((((((...))))..........(((-(((.....)))))).....))))))). ( -28.20, z-score = -2.09, R) >droSim1.chr3L 19273067 102 + 22553184 ---------------UCUUCUCAGGCAAUUUAUGACAUGCUAGGAGCGUGCUCAUCUAAUCGUCCAGCUGAUUCAGCAGAAGAAAGAGCA-AAAAAUAUUUUUGCAAAAAUGGACGAA ---------------................(((((((((.....))))).))))....(((((((((((...))))..........(((-(((.....)))))).....))))))). ( -28.20, z-score = -2.09, R) >droSec1.super_29 633646 102 + 700605 ---------------UCUUCUCAGGCAAUUUAUGACAUGCUAGGAGCGUGCUCAUCUAAUCGUCCAGCUGAUUCAGCAGAAGAAAGAGCA-AAAAAUAUUUUUGCAAAAAUGGACGAA ---------------................(((((((((.....))))).))))....(((((((((((...))))..........(((-(((.....)))))).....))))))). ( -28.20, z-score = -2.09, R) >droWil1.scaffold_180727 938089 102 + 2741493 ---------------UUUUUACAGGCAAUUUACGAUAUGCUAGGUGCAUGCUCAUCUAAUCGUCCAGCUGAUUCUGCAGAAGAGCGAGCA-AAAAAUAUUUUUGCUAAAAUGGAUGAA ---------------..................(((((((.....))))).))......((((((((((..(((....))).))).((((-(((.....)))))))....))))))). ( -25.70, z-score = -1.21, R) >droGri2.scaffold_15110 615749 102 - 24565398 ---------------UCGUUUUAGGCAAUCUACGACAUGCUAGGAGCAUGCUCAUCAAAUCGUCCUGCUGACUCUGCAGAGGAGCGUGCA-AAAAAUAUAUUUGCUAAGAUGGAUGAA ---------------(((((((((.(((((((........)))).((((((((...........((((.......))))..)))))))).-..........))))))))))))..... ( -26.32, z-score = -0.47, R) >droMoj3.scaffold_6680 1484471 97 - 24764193 --------------------UCAGGCAAUCUAUGACAUGCUAGGCGCAUGCUCAUCAAAUCGUCCCGCUGAUUCAGCUGAGGAGCGAGCA-AAAAAUAUAUUUGCUAAGAUGGAUGAA --------------------...........(((((((((.....))))).))))....((((((((((..(((....))).))))((((-((.......)))))).....)))))). ( -26.60, z-score = -0.64, R) >droYak2.chr3L 3617176 102 - 24197627 ---------------UUCUCUCAGGCAAUUUAUGACAUGCUAGGAGCGUGCUCAUCUAAUCGUCCAGCUGAUUCAGCAGAAGAAAGAGCA-AAAAAUAUUUUUGCAAAAAUGGACGAA ---------------................(((((((((.....))))).))))....(((((((((((...))))..........(((-(((.....)))))).....))))))). ( -28.20, z-score = -2.09, R) >droEre2.scaffold_4784 19727794 102 + 25762168 ---------------UUCUCUCAGGCAAUUUAUGACAUGCUAGGAGCGUGCUCAUCUAAUCGUCCAGCUGAUUCAGCAGAAGAAAGAGCA-AAAAAUAUUUUUGCAAAAAUGGACGAA ---------------................(((((((((.....))))).))))....(((((((((((...))))..........(((-(((.....)))))).....))))))). ( -28.20, z-score = -2.09, R) >droAna3.scaffold_13337 4009978 99 - 23293914 ------------------UUUUAGGCAAUUUAUGACAUGCUAGGAGCGUGCUCAUCUAAUCGUCCAGCUGACUCAGCAGAAGAAAGAGCA-AAAAAUAUUUUUGCAAAAAUGGACGAA ------------------.............(((((((((.....))))).))))....(((((((((((...))))..........(((-(((.....)))))).....))))))). ( -28.20, z-score = -2.69, R) >droPer1.super_88 185418 104 - 213009 -------------UAUUGUUACAGGCAAUUUAUGAUAUGCUUGGAGCGUGCUCAUCCAAUCGUCCUGCAGACUCCGCGGAAGAACGAGCA-AAAAAUAUAUUUGCUAAGAUGGACGAA -------------..(((((...(((((..(((....(((((((((....)))......((.(((.((.......))))).)).))))))-....)))...)))))......))))). ( -24.30, z-score = -0.44, R) >apiMel3.Group3 8692246 117 + 12341916 ACACUUUAAAUCGAUUUGUUUCAGGAUGCCGGUCUCGAGAGAAAGGAUUACCUUACGUAUAACGAUUUCAAGUUGAU-GAUGAAAGAGUACAAAGGCGAUUUCGUGGCAAUCGGUCUU .........(((((((.((..(..((((((.(((((......((((....)))).(((.((((........))))))-)......))).))...))).)))..)..)))))))))... ( -25.10, z-score = 0.47, R) >droVir3.scaffold_13049 7312017 102 - 25233164 ---------------UUUUUUCAGGCAAUCUAUGACAUGCUAGGAGCAUGCUCAUCAAAUCGUCCCGCUGAUUCUGCAGAGGAGCGUGCA-AAAAAUAUAUUUGCUAAAAUGGAUGAA ---------------................(((((((((.....))))).))))....((((((((((..(((....))).)))).(((-((.......)))))......)))))). ( -24.80, z-score = -0.38, R) >anoGam1.chr2L 44041558 102 + 48795086 ---------------UCUUCGCAGGCAAUAUACGACAUGCUGGGCGCGUGCUCAUCGAACAAACCGGCCGACUCGGCGGAGGAGCGCGCG-AAAAACAUCUUCGCCAAGAUGGAUGAA ---------------..(((((.((((..........))))..))((((((((..((.......))((((...))))....)))))))))-))...(((((((.....)).))))).. ( -33.00, z-score = -0.43, R) >triCas2.chrUn_22 88489 102 - 327011 ---------------UUUUUCCAGGCUAUUUACGACAUGUUGGGCGCCUGUUCGUCCAACCGCCCUGCGGAUUCCGCCGAAGAGCGGGCA-AAGAACAUCUUCGCGAAAAUGGACGAG ---------------......(((((......(((....)))...))))).(((((((.((((.((.(((......))).)).))))((.-(((.....))).)).....))))))). ( -35.60, z-score = -1.67, R) >consensus _______________UUUUUUCAGGCAAUUUAUGACAUGCUAGGAGCGUGCUCAUCUAAUCGUCCAGCUGAUUCAGCAGAAGAAAGAGCA_AAAAAUAUUUUUGCAAAAAUGGACGAA .................................(((((((.....))))).)).......(((((.((((...))))..........(((............)))......))))).. (-13.72 = -13.33 + -0.39)


| Location | 19,988,746 – 19,988,836 |
|---|---|
| Length | 90 |
| Sequences | 13 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 86.31 |
| Shannon entropy | 0.31107 |
| G+C content | 0.43162 |
| Mean single sequence MFE | -23.31 |
| Consensus MFE | -16.74 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.572249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
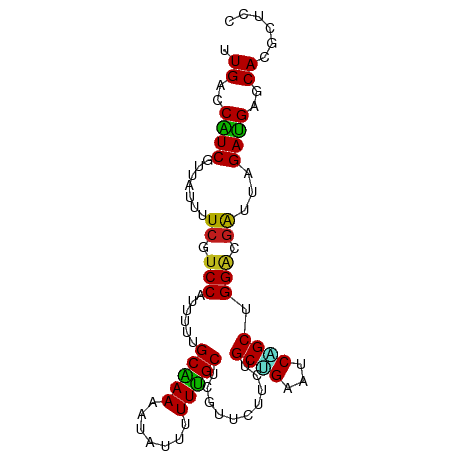
>dm3.chr3L 19988746 90 + 24543557 GGAGCGUGCUCAUCUAAUCGUCCAGCUGAUUCAGCAGAAGAAAGAGCAAAAAAUAUUUUUGCAAAAAUGGACGAAAAUAACGAUGGUCAA .(((....)))......(((((((((((...))))..........((((((.....)))))).....)))))))................ ( -23.00, z-score = -2.30, R) >droSim1.chr3L 19273094 90 + 22553184 GGAGCGUGCUCAUCUAAUCGUCCAGCUGAUUCAGCAGAAGAAAGAGCAAAAAAUAUUUUUGCAAAAAUGGACGAAAAUAACGAUGGUCAA .(((....)))......(((((((((((...))))..........((((((.....)))))).....)))))))................ ( -23.00, z-score = -2.30, R) >droSec1.super_29 633673 90 + 700605 GGAGCGUGCUCAUCUAAUCGUCCAGCUGAUUCAGCAGAAGAAAGAGCAAAAAAUAUUUUUGCAAAAAUGGACGAAAAUAACGAUGGUCAA .(((....)))......(((((((((((...))))..........((((((.....)))))).....)))))))................ ( -23.00, z-score = -2.30, R) >droWil1.scaffold_180727 938116 90 + 2741493 GGUGCAUGCUCAUCUAAUCGUCCAGCUGAUUCUGCAGAAGAGCGAGCAAAAAAUAUUUUUGCUAAAAUGGAUGAAAAUAAUGAUGGUCAA ......((..((((...((((((((((..(((....))).))).(((((((.....)))))))....))))))).......))))..)). ( -23.40, z-score = -1.76, R) >droGri2.scaffold_15110 615776 90 - 24565398 GGAGCAUGCUCAUCAAAUCGUCCUGCUGACUCUGCAGAGGAGCGUGCAAAAAAUAUAUUUGCUAAGAUGGAUGAAAAUAAUGAUGGUCAA ......((..(((((..((((((.(((..(((....))).)))..(((((.......)))))......))))))......)))))..)). ( -21.80, z-score = -0.81, R) >droMoj3.scaffold_6680 1484493 90 - 24764193 GGCGCAUGCUCAUCAAAUCGUCCCGCUGAUUCAGCUGAGGAGCGAGCAAAAAAUAUAUUUGCUAAGAUGGAUGAAAAUAACGAUGGUCAA ......((..((((...((((((((((..(((....))).))))((((((.......)))))).....)))))).......))))..)). ( -21.30, z-score = -0.33, R) >droYak2.chr3L 3617203 90 - 24197627 GGAGCGUGCUCAUCUAAUCGUCCAGCUGAUUCAGCAGAAGAAAGAGCAAAAAAUAUUUUUGCAAAAAUGGACGAAAAUAACGAUGGUCAA .(((....)))......(((((((((((...))))..........((((((.....)))))).....)))))))................ ( -23.00, z-score = -2.30, R) >droEre2.scaffold_4784 19727821 90 + 25762168 GGAGCGUGCUCAUCUAAUCGUCCAGCUGAUUCAGCAGAAGAAAGAGCAAAAAAUAUUUUUGCAAAAAUGGACGAAAAUAACGAUGGUCAA .(((....)))......(((((((((((...))))..........((((((.....)))))).....)))))))................ ( -23.00, z-score = -2.30, R) >droAna3.scaffold_13337 4010002 90 - 23293914 GGAGCGUGCUCAUCUAAUCGUCCAGCUGACUCAGCAGAAGAAAGAGCAAAAAAUAUUUUUGCAAAAAUGGACGAAAAUAACGAUGGUCAA .(((....)))......(((((((((((...))))..........((((((.....)))))).....)))))))................ ( -23.00, z-score = -2.28, R) >droPer1.super_88 185447 90 - 213009 GGAGCGUGCUCAUCCAAUCGUCCUGCAGACUCCGCGGAAGAACGAGCAAAAAAUAUAUUUGCUAAGAUGGACGAAAAUAAUGAUGGUCAA .(((....)))......((((((...........(....)....((((((.......)))))).....))))))................ ( -18.20, z-score = 0.21, R) >droVir3.scaffold_13049 7312044 90 - 25233164 GGAGCAUGCUCAUCAAAUCGUCCCGCUGAUUCUGCAGAGGAGCGUGCAAAAAAUAUAUUUGCUAAAAUGGAUGAAAAUAACGACGGUCAA .(((....)))......((((((((((..(((....))).)))).(((((.......)))))......))))))................ ( -19.20, z-score = -0.16, R) >anoGam1.chr2L 44041585 90 + 48795086 GGCGCGUGCUCAUCGAACAAACCGGCCGACUCGGCGGAGGAGCGCGCGAAAAACAUCUUCGCCAAGAUGGAUGAAAAUAACGACGGUCAG (((((((((((..((.......))((((...))))....))))))))(((.......))))))..(((.(.((.......)).).))).. ( -29.10, z-score = -0.87, R) >triCas2.chrUn_22 88516 90 - 327011 GGCGCCUGUUCGUCCAACCGCCCUGCGGAUUCCGCCGAAGAGCGGGCAAAGAACAUCUUCGCGAAAAUGGACGAGAACAACGACGGCCAA (((.....((((((((.((((.((.(((......))).)).))))((.(((.....))).)).....))))))))..(......)))).. ( -32.00, z-score = -1.37, R) >consensus GGAGCGUGCUCAUCUAAUCGUCCAGCUGAUUCAGCAGAAGAACGAGCAAAAAAUAUUUUUGCAAAAAUGGACGAAAAUAACGAUGGUCAA ......((..((((...((((((.((((...))))..........(((((.......)))))......)))))).......))))..)). (-16.74 = -16.93 + 0.19)
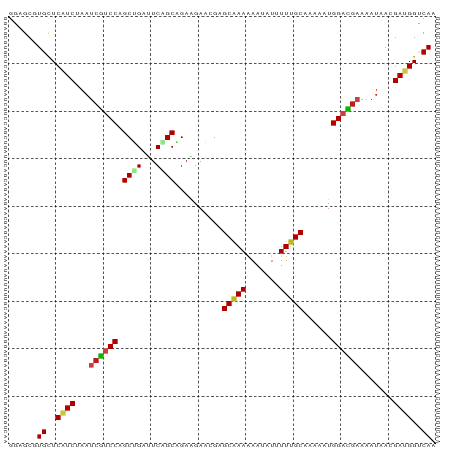
| Location | 19,988,746 – 19,988,836 |
|---|---|
| Length | 90 |
| Sequences | 13 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 86.31 |
| Shannon entropy | 0.31107 |
| G+C content | 0.43162 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -14.44 |
| Energy contribution | -13.90 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.540366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
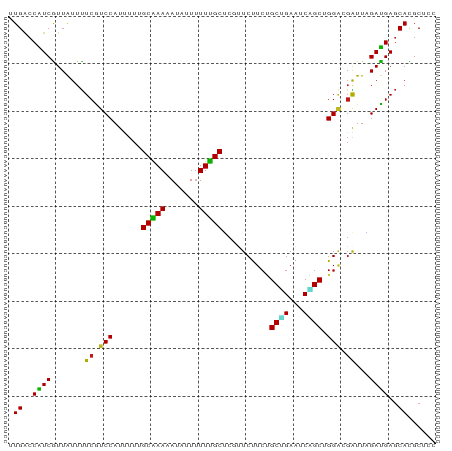
>dm3.chr3L 19988746 90 - 24543557 UUGACCAUCGUUAUUUUCGUCCAUUUUUGCAAAAAUAUUUUUUGCUCUUUCUUCUGCUGAAUCAGCUGGACGAUUAGAUGAGCACGCUCC .((..((((.......(((((((.....((((((.....))))))..........((((...)))))))))))...))))..))...... ( -22.30, z-score = -2.78, R) >droSim1.chr3L 19273094 90 - 22553184 UUGACCAUCGUUAUUUUCGUCCAUUUUUGCAAAAAUAUUUUUUGCUCUUUCUUCUGCUGAAUCAGCUGGACGAUUAGAUGAGCACGCUCC .((..((((.......(((((((.....((((((.....))))))..........((((...)))))))))))...))))..))...... ( -22.30, z-score = -2.78, R) >droSec1.super_29 633673 90 - 700605 UUGACCAUCGUUAUUUUCGUCCAUUUUUGCAAAAAUAUUUUUUGCUCUUUCUUCUGCUGAAUCAGCUGGACGAUUAGAUGAGCACGCUCC .((..((((.......(((((((.....((((((.....))))))..........((((...)))))))))))...))))..))...... ( -22.30, z-score = -2.78, R) >droWil1.scaffold_180727 938116 90 - 2741493 UUGACCAUCAUUAUUUUCAUCCAUUUUAGCAAAAAUAUUUUUUGCUCGCUCUUCUGCAGAAUCAGCUGGACGAUUAGAUGAGCAUGCACC .((....(((((....((.((((....(((((((.....))))))).(((.(((....)))..))))))).))...))))).....)).. ( -16.90, z-score = -0.73, R) >droGri2.scaffold_15110 615776 90 + 24565398 UUGACCAUCAUUAUUUUCAUCCAUCUUAGCAAAUAUAUUUUUUGCACGCUCCUCUGCAGAGUCAGCAGGACGAUUUGAUGAGCAUGCUCC .......((((((...((.(((......(((((.......)))))..(((.(((....)))..))).))).))..))))))......... ( -20.30, z-score = -1.55, R) >droMoj3.scaffold_6680 1484493 90 + 24764193 UUGACCAUCGUUAUUUUCAUCCAUCUUAGCAAAUAUAUUUUUUGCUCGCUCCUCAGCUGAAUCAGCGGGACGAUUUGAUGAGCAUGCGCC ........(((.((.((((((.......(((((.......)))))(((.((((..((((...)))))))))))...)))))).))))).. ( -20.50, z-score = -0.78, R) >droYak2.chr3L 3617203 90 + 24197627 UUGACCAUCGUUAUUUUCGUCCAUUUUUGCAAAAAUAUUUUUUGCUCUUUCUUCUGCUGAAUCAGCUGGACGAUUAGAUGAGCACGCUCC .((..((((.......(((((((.....((((((.....))))))..........((((...)))))))))))...))))..))...... ( -22.30, z-score = -2.78, R) >droEre2.scaffold_4784 19727821 90 - 25762168 UUGACCAUCGUUAUUUUCGUCCAUUUUUGCAAAAAUAUUUUUUGCUCUUUCUUCUGCUGAAUCAGCUGGACGAUUAGAUGAGCACGCUCC .((..((((.......(((((((.....((((((.....))))))..........((((...)))))))))))...))))..))...... ( -22.30, z-score = -2.78, R) >droAna3.scaffold_13337 4010002 90 + 23293914 UUGACCAUCGUUAUUUUCGUCCAUUUUUGCAAAAAUAUUUUUUGCUCUUUCUUCUGCUGAGUCAGCUGGACGAUUAGAUGAGCACGCUCC .((..((((.......(((((((.....((((((.....))))))..........((((...)))))))))))...))))..))...... ( -22.30, z-score = -2.36, R) >droPer1.super_88 185447 90 + 213009 UUGACCAUCAUUAUUUUCGUCCAUCUUAGCAAAUAUAUUUUUUGCUCGUUCUUCCGCGGAGUCUGCAGGACGAUUGGAUGAGCACGCUCC ...............((((((((.....(((((.......)))))(((((((...((((...))))))))))).))))))))........ ( -23.00, z-score = -1.63, R) >droVir3.scaffold_13049 7312044 90 + 25233164 UUGACCGUCGUUAUUUUCAUCCAUUUUAGCAAAUAUAUUUUUUGCACGCUCCUCUGCAGAAUCAGCGGGACGAUUUGAUGAGCAUGCUCC .......((((((...((.(((......(((((.......))))).((((..((....))...))))))).))..))))))......... ( -18.70, z-score = -0.58, R) >anoGam1.chr2L 44041585 90 - 48795086 CUGACCGUCGUUAUUUUCAUCCAUCUUGGCGAAGAUGUUUUUCGCGCGCUCCUCCGCCGAGUCGGCCGGUUUGUUCGAUGAGCACGCGCC ...........(((((((..((.....)).))))))).....((((.((((....((((...))))(((.....)))..)))).)))).. ( -23.40, z-score = 1.08, R) >triCas2.chrUn_22 88516 90 + 327011 UUGGCCGUCGUUGUUCUCGUCCAUUUUCGCGAAGAUGUUCUUUGCCCGCUCUUCGGCGGAAUCCGCAGGGCGGUUGGACGAACAGGCGCC ..(((.(((..(((..(((((((.....((((((.....))))))(((((((.(((......))).))))))).)))))))))))))))) ( -39.10, z-score = -2.28, R) >consensus UUGACCAUCGUUAUUUUCGUCCAUUUUUGCAAAAAUAUUUUUUGCUCGUUCUUCUGCUGAAUCAGCUGGACGAUUAGAUGAGCACGCUCC .((..((((.......((.(((......(((((.......)))))..........((((...)))).))).))...))))..))...... (-14.44 = -13.90 + -0.54)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:51 2011