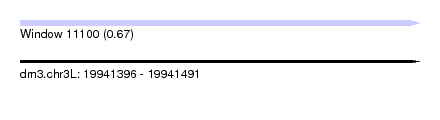
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,941,396 – 19,941,491 |
| Length | 95 |
| Max. P | 0.671170 |

| Location | 19,941,396 – 19,941,491 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 63.68 |
| Shannon entropy | 0.71836 |
| G+C content | 0.42740 |
| Mean single sequence MFE | -23.01 |
| Consensus MFE | -6.59 |
| Energy contribution | -7.34 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.671170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19941396 95 + 24543557 UCGCAUGCCUUAAGGAUAUGCGA--GAACUUCAGUCGAGGCGGCUUGACA----UACUGACAUUUAUUACACAGUGCACUUUUGGUCCAGCAAUGUACUCG-- (((((((((....)).)))))))--..........((((((.(((.(((.----(((((............)))))..(....)))).)))...)).))))-- ( -22.80, z-score = -0.52, R) >droPer1.super_20 204879 93 - 1872136 UCGCAUGCCUGCAGGAUAUGCGA--CAACUUUGCUUGAGUUGUCCCGACA----UACUGACAUUUGUU-UAAGGAUUGUUUAGGGCUUGUUGAGGACUUG--- (((((((((....)).)))))))--.(((((.....)))))(((((((((----..(((((((((...-...))).)).))))....))))).))))...--- ( -25.80, z-score = -1.47, R) >dp4.chrXR_group6 1570827 93 - 13314419 UCGCAUGCCUGCAGGAUAUGCGA--CAACUUUGCUUGAGUUGCCCCGACA----UACUGACAUUUGUU-UAAGGAUUGUUUAGGGCUUGUUGAGGACUUG--- (((((((((....)).)))))))--........((..(...((((.((((----..(((((....)))-..))...))))..))))...)..))......--- ( -24.10, z-score = -0.92, R) >droAna3.scaffold_13337 3967366 88 - 23293914 UCGCAUUCCUUAAGGAAAUGCGA--GAACUUCACUCGAGUUGGAUAGACA----UACUGAUGUUUGUG-CAAAGGACAUGUCCUACAGAGUCUGG-------- (((((((((....)).)))))))--...((..((((...(((.(((((((----(....)))))))).-)))((((....))))...))))..))-------- ( -28.70, z-score = -2.88, R) >droEre2.scaffold_4784 19679508 89 + 25762168 UCGCAUGCCUUCGGGAUAUGCGA--GAACUUCAAGCGA-----CUUGACA----UACUGACAUUUAUUACACAGUACACUUUUG-UCUAGCAAUUUACUUG-- (((((((((....)).)))))))--.........((..-----...((((----(((((............)))))......))-))..))..........-- ( -20.70, z-score = -2.95, R) >droSec1.super_29 586126 95 + 700605 UCGCAUGCCUUAAGUAUAUGCGA--GAACUUCAGUCGAGCCGGCUUGACA----UACUGACAUUUAUUACACAGUACACUUUUGGUCUUCCAAUUUACUCG-- (((((((.........)))))))--........((((((....)))))).----(((((............))))).....((((....))))........-- ( -19.40, z-score = -1.35, R) >droSim1.chrX 5096484 94 - 17042790 UCGCAUGCCUUAAGGAUAUGCGA--GAACUUCAGUCGAGCCGGCUUGACA----UACUGACAUUUAUUACACAGUACACUUUUGGUCCACCAAUUU-CUCG-- ..(((((((....)).)))))((--(((.....((((((....)))))).----(((((............))))).....((((....)))).))-))).-- ( -26.10, z-score = -3.46, R) >triCas2.ChLG7 7255835 98 - 17478683 ----ACGGAGCGAGU-CAUGCGAUCGACGUUUUGUUGGCUAAGCUUGACAAUUUUAUUACAAUUUAUUUCAAAGCUUUUUUGUAAAAUACUGAAUAAAUUGGU ----...((((.(((-((.((((........)))))))))..)))).............((((((((((...((..(((.....)))..)))))))))))).. ( -16.50, z-score = 0.33, R) >consensus UCGCAUGCCUUAAGGAUAUGCGA__GAACUUCAGUCGAGUCGGCUUGACA____UACUGACAUUUAUUACAAAGUACACUUUUGGUCUACCAAUGUACUCG__ (((((((((....)).)))))))................................................................................ ( -6.59 = -7.34 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:47 2011