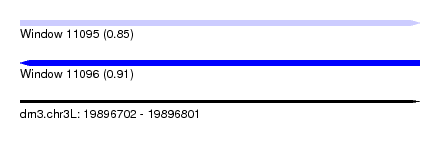
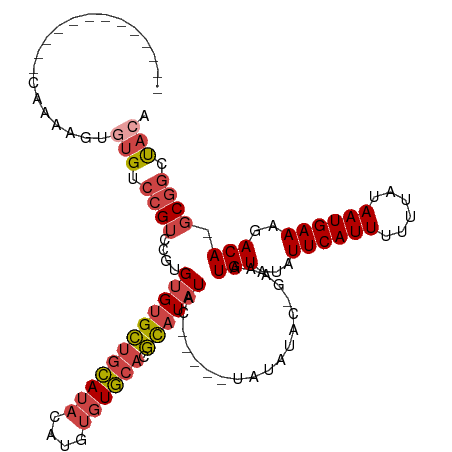
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,896,702 – 19,896,801 |
| Length | 99 |
| Max. P | 0.913070 |

| Location | 19,896,702 – 19,896,801 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.05 |
| Shannon entropy | 0.32960 |
| G+C content | 0.37265 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -13.59 |
| Energy contribution | -14.37 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.848212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19896702 99 + 24543557 UGUAGCCGC-UGUCUUUCAUUAUAAAAAAUGAAUAUACAUUUC-GUAUAUA-----GUAUAUGCGUGCACACAUGUAUGCAGCACACACGGACGGACAAACUUUUC------------- ....(((((-(((...(((((......)))))((((((.....-)))))).-----((...((((((((....))))))))..))..)))).))).).........------------- ( -22.80, z-score = -1.45, R) >droEre2.scaffold_4784 19634994 97 + 25762168 UGUAGCCGC-UGUCUUUCAUUAUAAAAAAUGAAUAUACAUUUC-GUAUAUA-----GUAUAUGCGUGCACACAUGUAUGCAGCACAC--GGACGGACACACUUUUG------------- (((..((((-(((...(((((......)))))((((((.....-)))))).-----((...((((((((....))))))))..))))--)).)))..)))......------------- ( -24.90, z-score = -1.85, R) >droYak2.chr3L 3517286 97 - 24197627 UGUAGCCGC-UGUCUUUCAUUAUAAAAAAUGAAUAUACAUUAC-GUAUAUA-----GUAUAUGCGUGCACACAUGUAUGCAGCACAC--GGACGGACACACUUUUA------------- (((..((((-(((...(((((......)))))((((((.....-)))))).-----((...((((((((....))))))))..))))--)).)))..)))......------------- ( -24.90, z-score = -1.85, R) >droSec1.super_29 542078 99 + 700605 UGUAGCCGC-UGUCUUUCAUUAUAAAAAAUGAAUAUACAUUUC-GUAUAUA-----GUAUAUGCGUGCACACAUGUAUGCAGCACACACGGACGGACACACUUUUG------------- (((..((((-(((...(((((......)))))((((((.....-)))))).-----((...((((((((....))))))))..))..)))).)))..)))......------------- ( -24.90, z-score = -1.75, R) >droSim1.chr3L 19191352 99 + 22553184 UGUAGCCGC-UGUCUUUCAUUAUAAAAAAUGAAUAUACAUUUC-GUAUAUA-----GUAUAUGCGUGCACACAUGUAUGCAGCACACACGGACGGACACACUUUUG------------- (((..((((-(((...(((((......)))))((((((.....-)))))).-----((...((((((((....))))))))..))..)))).)))..)))......------------- ( -24.90, z-score = -1.75, R) >droWil1.scaffold_180698 2809688 117 - 11422946 --UGACGGCAUGUCUUUCAUUAUACAAAAUGAAUAUACAUUUUUGUAUAUACAUUUGUAUAUAUCUUUACCCCCCUCUAUCACCCUCUCCCUCUCACUGACUAUGUCGAUCUCUUUUUU --.(((((((((((......((((((((.((.(((((((....))))))).))))))))))((....)).............................))).)))))).))........ ( -21.20, z-score = -5.10, R) >consensus UGUAGCCGC_UGUCUUUCAUUAUAAAAAAUGAAUAUACAUUUC_GUAUAUA_____GUAUAUGCGUGCACACAUGUAUGCAGCACACACGGACGGACACACUUUUG_____________ ...........((((.(((((......)))))((((((......))))))......((...(((((((......)))))))..))........))))...................... (-13.59 = -14.37 + 0.78)

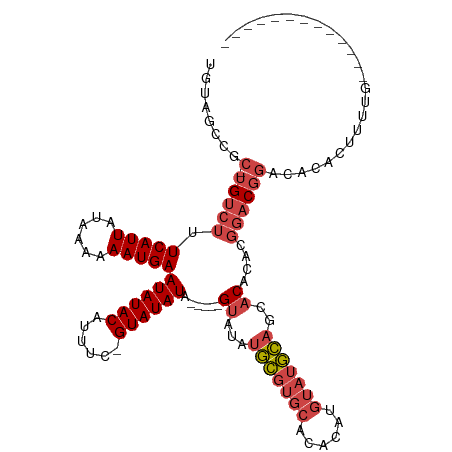
| Location | 19,896,702 – 19,896,801 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.05 |
| Shannon entropy | 0.32960 |
| G+C content | 0.37265 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -18.69 |
| Energy contribution | -19.33 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19896702 99 - 24543557 -------------GAAAAGUUUGUCCGUCCGUGUGUGCUGCAUACAUGUGUGCACGCAUAUAC-----UAUAUAC-GAAAUGUAUAUUCAUUUUUUAUAAUGAAAGACA-GCGGCUACA -------------....((((((((..((.((((((((((((((....)))))).))))))))-----..((((.-((((((......)))))).))))..))..))))-)..)))... ( -29.70, z-score = -2.67, R) >droEre2.scaffold_4784 19634994 97 - 25762168 -------------CAAAAGUGUGUCCGUCC--GUGUGCUGCAUACAUGUGUGCACGCAUAUAC-----UAUAUAC-GAAAUGUAUAUUCAUUUUUUAUAAUGAAAGACA-GCGGCUACA -------------......((((.((((..--((((((((((((....)))))).))))))..-----.......-....(((...((((((......))))))..)))-)))).)))) ( -26.50, z-score = -1.53, R) >droYak2.chr3L 3517286 97 + 24197627 -------------UAAAAGUGUGUCCGUCC--GUGUGCUGCAUACAUGUGUGCACGCAUAUAC-----UAUAUAC-GUAAUGUAUAUUCAUUUUUUAUAAUGAAAGACA-GCGGCUACA -------------......((((.((((..--((((((((((((....)))))).))))))..-----.......-....(((...((((((......))))))..)))-)))).)))) ( -26.50, z-score = -1.33, R) >droSec1.super_29 542078 99 - 700605 -------------CAAAAGUGUGUCCGUCCGUGUGUGCUGCAUACAUGUGUGCACGCAUAUAC-----UAUAUAC-GAAAUGUAUAUUCAUUUUUUAUAAUGAAAGACA-GCGGCUACA -------------......((((.((((..((((((((((((((....)))))).))))))))-----.......-....(((...((((((......))))))..)))-)))).)))) ( -30.70, z-score = -2.50, R) >droSim1.chr3L 19191352 99 - 22553184 -------------CAAAAGUGUGUCCGUCCGUGUGUGCUGCAUACAUGUGUGCACGCAUAUAC-----UAUAUAC-GAAAUGUAUAUUCAUUUUUUAUAAUGAAAGACA-GCGGCUACA -------------......((((.((((..((((((((((((((....)))))).))))))))-----.......-....(((...((((((......))))))..)))-)))).)))) ( -30.70, z-score = -2.50, R) >droWil1.scaffold_180698 2809688 117 + 11422946 AAAAAAGAGAUCGACAUAGUCAGUGAGAGGGAGAGGGUGAUAGAGGGGGGUAAAGAUAUAUACAAAUGUAUAUACAAAAAUGUAUAUUCAUUUUGUAUAAUGAAAGACAUGCCGUCA-- ............(((...((((.(...........).))))...((.(.((.......((((((((((.(((((((....))))))).)).)))))))).......)).).))))).-- ( -20.54, z-score = -1.88, R) >consensus _____________CAAAAGUGUGUCCGUCCGUGUGUGCUGCAUACAUGUGUGCACGCAUAUAC_____UAUAUAC_GAAAUGUAUAUUCAUUUUUUAUAAUGAAAGACA_GCGGCUACA ..................((.((((.......((((((((((((....)))))).)))))).........(((((......)))))((((((......)))))).)))).))....... (-18.69 = -19.33 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:43 2011