
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,883,225 – 19,883,339 |
| Length | 114 |
| Max. P | 0.865123 |

| Location | 19,883,225 – 19,883,339 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 143 |
| Reading direction | forward |
| Mean pairwise identity | 63.12 |
| Shannon entropy | 0.52962 |
| G+C content | 0.39717 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -9.74 |
| Energy contribution | -11.02 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.865123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
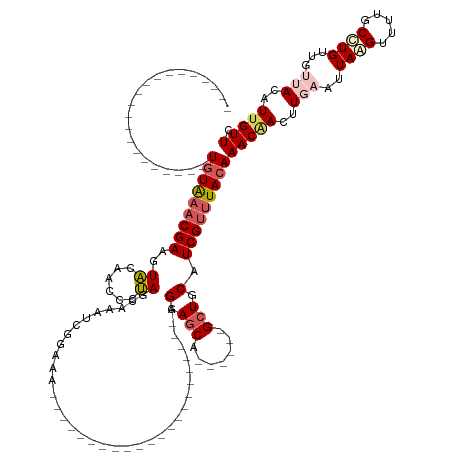
>dm3.chr3L 19883225 114 + 24543557 UGUAAACGAUGCAACGGCAACUAUAC-CGAAGUACAAGCAUAGAAAAUCGUUUA----------------------AGGAUCA------GCUCCAUCGUUUACAAACCACUUGAAUUAAGUUUUGCUUGUUGUUACAUUGUUU ...((((((((.((((....).....-......(((((((.(((.....(((((----------------------(((((..------.....)))(((....)))..)))))))....)))))))))).))).)))))))) ( -21.00, z-score = -0.14, R) >droSim1.chr3L 19177832 94 + 22553184 ---------------------UGUAAACGAAGUACAAUCCUAGCAAAUCGCCCA----------------------AGGAGCA------GCUGCAUCGUUUACAAACAACUUGAAUUAAGUUUUGCCUGUUGUUACAUUGUUG ---------------------(((((((((.((((..((((.((.....))...----------------------))))...------).))).)))))))))((((((.((((......))))...))))))......... ( -21.00, z-score = -1.24, R) >droSec1.super_29 528540 94 + 700605 ---------------------UGUAAACGAAGUACAAUCCUAGCAAAUCGGCCA----------------------AGGAGCA------GCUGCAUCGUUUACAAACAACUUGAAUUAAGUUUUGCCUGUUGUUACAUUGUUU ---------------------(((((((((.((((..((((.((......))..----------------------))))...------).))).)))))))))((((((.((((......))))...))))))......... ( -21.60, z-score = -1.12, R) >droYak2.chr3L 3503550 117 - 24197627 ---------------------UGUGAACGA--UGAAGCGGCAACAAGUCCGAAAUUGAAGCCUAAGAAAUCGGCUCAGGAGCUUAUAUCGCUUCAUCGUUUACAAACAACUUGAAUUAGGUUUUGCCUGUU---AUAUUGUUC ---------------------(((((((((--(((((((((..(((........)))..)))...((.((.((((....)))).)).)))))))))))))))))(((((..(((..((((.....))))))---)..))))). ( -41.70, z-score = -5.24, R) >droEre2.scaffold_4784 19621389 96 + 25762168 ---------------------UGUAAACGA--UGCAGCGGCAGCGAGUCCAAAGU--------------------CAGGAGCUUACAUCGCU-CAUCGUUUACAAACGAUGUGAAUUAGGUUUGACCUGUU---AUGUUGUUC ---------------------(((((((((--((.(((((..(..(((((.....--------------------..))).))..).)))))-)))))))))))((((((((((..((((.....))))))---)))))))). ( -35.10, z-score = -4.00, R) >consensus _____________________UGUAAACGAAGUACAACCCUAGCAAAUCGGAAA______________________AGGAGCA______GCUGCAUCGUUUACAAACAACUUGAAUUAAGUUUUGCCUGUUGUUACAUUGUUC .....................(((((((((..((......))...................................(.(((.......))).).)))))))))(((((..(((..((((.....))))...)))..))))). ( -9.74 = -11.02 + 1.28)

| Location | 19,883,225 – 19,883,339 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 143 |
| Reading direction | reverse |
| Mean pairwise identity | 63.12 |
| Shannon entropy | 0.52962 |
| G+C content | 0.39717 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -8.38 |
| Energy contribution | -8.34 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19883225 114 - 24543557 AAACAAUGUAACAACAAGCAAAACUUAAUUCAAGUGGUUUGUAAACGAUGGAGC------UGAUCCU----------------------UAAACGAUUUUCUAUGCUUGUACUUCG-GUAUAGUUGCCGUUGCAUCGUUUACA ((((.(((((((.((((((...((((.....)))).)))))).(((.((((((.------.((((..----------------------.....))))))))))...(((((....-))))))))...))))))).))))... ( -28.10, z-score = -2.01, R) >droSim1.chr3L 19177832 94 - 22553184 CAACAAUGUAACAACAGGCAAAACUUAAUUCAAGUUGUUUGUAAACGAUGCAGC------UGCUCCU----------------------UGGGCGAUUUGCUAGGAUUGUACUUCGUUUACA--------------------- .((((((........((......))........))))))((((((((((((((.------...((((----------------------..(((.....))))))))))))..)))))))))--------------------- ( -20.09, z-score = -0.17, R) >droSec1.super_29 528540 94 - 700605 AAACAAUGUAACAACAGGCAAAACUUAAUUCAAGUUGUUUGUAAACGAUGCAGC------UGCUCCU----------------------UGGCCGAUUUGCUAGGAUUGUACUUCGUUUACA--------------------- (((((((........((......))........)))))))(((((((((((((.------...((((----------------------.(((......))))))))))))..)))))))).--------------------- ( -22.09, z-score = -1.09, R) >droYak2.chr3L 3503550 117 + 24197627 GAACAAUAU---AACAGGCAAAACCUAAUUCAAGUUGUUUGUAAACGAUGAAGCGAUAUAAGCUCCUGAGCCGAUUUCUUAGGCUUCAAUUUCGGACUUGUUGCCGCUUCA--UCGUUCACA--------------------- (((((((..---...(((.....))).......)))))))((.((((((((((((..(((((.(((((((((.........))).))).....))))))))...)))))))--))))).)).--------------------- ( -37.30, z-score = -4.62, R) >droEre2.scaffold_4784 19621389 96 - 25762168 GAACAACAU---AACAGGUCAAACCUAAUUCACAUCGUUUGUAAACGAUG-AGCGAUGUAAGCUCCUG--------------------ACUUUGGACUCGCUGCCGCUGCA--UCGUUUACA--------------------- .........---...(((.....))).............(((((((((((-((((..(..((.(((..--------------------.....)))))..)...)))).))--)))))))))--------------------- ( -28.20, z-score = -3.13, R) >consensus AAACAAUGUAACAACAGGCAAAACUUAAUUCAAGUUGUUUGUAAACGAUGCAGC______UGCUCCU______________________UGGCCGAUUUGCUACGAUUGUACUUCGUUUACA_____________________ (((((((........(((.....))).......)))))))(((((((((((((........((....................................)).....)))))..))))))))...................... ( -8.38 = -8.34 + -0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:40 2011