| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,773,552 – 5,773,619 |
| Length | 67 |
| Max. P | 0.859417 |

| Location | 5,773,552 – 5,773,619 |
|---|---|
| Length | 67 |
| Sequences | 11 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 77.37 |
| Shannon entropy | 0.47869 |
| G+C content | 0.35853 |
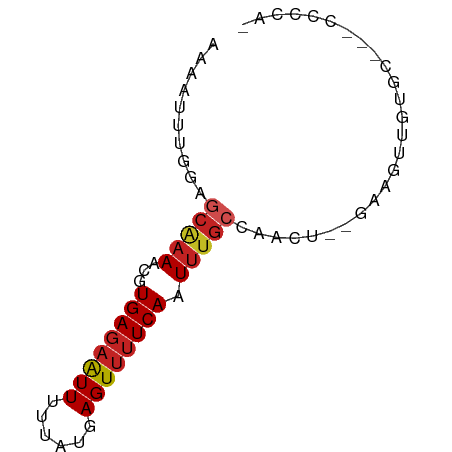
| Mean single sequence MFE | -12.70 |
| Consensus MFE | -7.12 |
| Energy contribution | -7.47 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859417 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
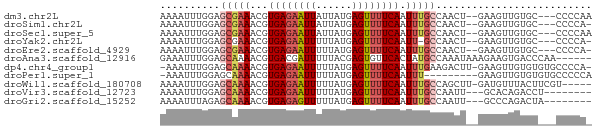
>dm3.chr2L 5773552 67 + 23011544 AAAAUUUGGAGCGAAACGUGAGAAUUAUUAUGAGUUUUCAAUUUGCCAACU--GAAGUUGUGC---CCCCAA .....((((.(((((...((((((((......)))))))).)))))((((.--...))))...---..)))) ( -14.50, z-score = -2.38, R) >droSim1.chr2L 5570281 66 + 22036055 AAAAUUUGGAGCGAAACGUGAGAAUUAUUAUGAGUUUUCAAUUUGCCAACU--GAAGUUGUGC---CCCCA- ......(((.(((((...((((((((......)))))))).)))))((((.--...))))...---..)))- ( -13.60, z-score = -2.02, R) >droSec1.super_5 3852389 67 + 5866729 AAAAUUUGGAGCGAAACGUGAGAAUUAUUAUGAGUUUUCAAUUUGCCAACU--GAAGUUGUGC---CCCCAA .....((((.(((((...((((((((......)))))))).)))))((((.--...))))...---..)))) ( -14.50, z-score = -2.38, R) >droYak2.chr2L 8906905 65 - 22324452 AAAAUUUGGAGCGAAACGUGAGAAUUUUUAUGAGUUUUCAAUU-GCCAACU--GAAGUUGUGC---CCCCA- ......(((.((((....(((((((((....))))))))).))-))((((.--...))))...---..)))- ( -13.40, z-score = -1.71, R) >droEre2.scaffold_4929 5865390 66 + 26641161 AAAAUUUGGAGCGAAACGUGAGAAUUUUUAUGAGUUUUCAAUUUGCCAACU--GAAGUUGUGC---CCCCA- ......(((.(((((...(((((((((....))))))))).)))))((((.--...))))...---..)))- ( -14.90, z-score = -2.29, R) >droAna3.scaffold_12916 11207491 66 + 16180835 GAAAUUUGGAGCAAAACGUGACGAUUUUUACGAGUGUUCACUAUGCCAAAUAAAGAAGUGACCCAA------ .....((((.(((...(((((......)))))..)))(((((..............))))).))))------ ( -9.74, z-score = -0.38, R) >dp4.chr4_group1 4666125 69 - 5278887 -AAAUUUGGAGCAAAACGUGAGAAUUUUUAUGAGUUUUCAAUUUGAAGACUU-GAAGUUGUGUGUGCCCCA- -.....(((.(((...(....)((((((...((((((((.....))))))))-)))))).....))).)))- ( -15.90, z-score = -2.35, R) >droPer1.super_1 9675325 62 - 10282868 -AAAUUUGGAGCAAAACGUGAGAAUUUUUAUGAGUUUUCAAUUU---------GAAGUUGUGUGUGCCCCCA -......((.(((..((((((((((((....)))))))))....---------.....)))...))).)).. ( -11.40, z-score = -1.40, R) >droWil1.scaffold_180708 4181296 66 - 12563649 AAAAUUUGGAGCAAAACGUGAGAAUUUUUAUGAGUUUUCAAUUUGCCAGCUU-GAUGUUUACUUCGU----- ...(((.((.(((((...(((((((((....))))))))).)))))...)).-)))...........----- ( -11.80, z-score = -0.57, R) >droVir3.scaffold_12723 1452358 61 - 5802038 AAAAUUUGGAGCAAAACGUGAGAAUUUUUAUGAGUUUUCAAUUUGCCAAUU---GCACAGACCU-------- ........(.(((((...(((((((((....))))))))).))))))....---..........-------- ( -11.30, z-score = -1.19, R) >droGri2.scaffold_15252 3335520 61 + 17193109 AAAAUUUAGAGCAAAACGUGAGAGUUUUUAUGAGUUUUCAAUUUGCCAAUU---GCCCAGACUA-------- ........(.(((((...(((((..((....))..))))).))))))....---..........-------- ( -8.70, z-score = -0.78, R) >consensus AAAAUUUGGAGCAAAACGUGAGAAUUUUUAUGAGUUUUCAAUUUGCCAACU__GAAGUUGUGC___CCCCA_ ..........(((((...((((((((......)))))))).))))).......................... ( -7.12 = -7.47 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:49 2011