| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,865,819 – 19,865,912 |
| Length | 93 |
| Max. P | 0.904022 |

| Location | 19,865,819 – 19,865,912 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 70.92 |
| Shannon entropy | 0.50156 |
| G+C content | 0.41609 |
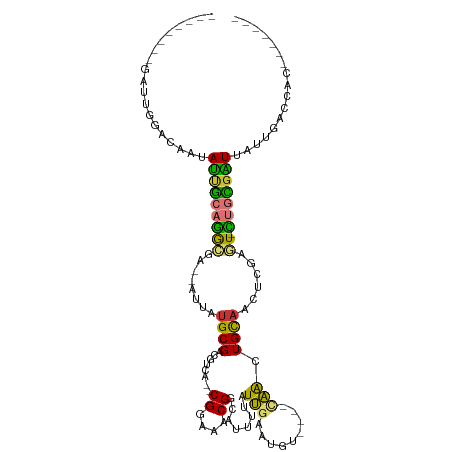
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -9.26 |
| Energy contribution | -8.46 |
| Covariance contribution | -0.79 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
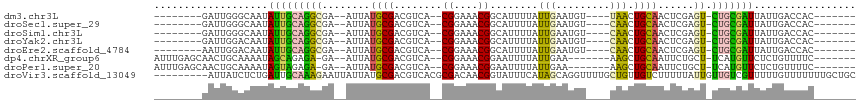
>dm3.chr3L 19865819 93 - 24543557 --------GAUUGGGCAAUAUUGCAGGCGA--AUUAUGCGACGUCA--CGGAAACGGCAUUUUAUUGAAUGU----UAACUGCAACUCGAGU-CUGCGAUUAUUGACCAC------- --------...(((.(((((((((((((..--....((((......--((....))((((((....))))))----....))))......))-)))))).))))).))).------- ( -29.70, z-score = -3.45, R) >droSec1.super_29 511220 93 - 700605 --------GAUUGGGCAAUAUUGCAGGCGA--AUUAUGCGACGUCA--CGGAAACGGCAUUUUAUUGAAUGU----CAACUGCAACUCGAGU-CUGCGAUUAUUGACCAC------- --------...(((.(((((((((((((..--....((((......--.(....)(((((((....))))))----)...))))......))-)))))).))))).))).------- ( -31.00, z-score = -3.55, R) >droSim1.chr3L 19157193 93 - 22553184 --------GAUUGGGCAAUAUUGCAGGCGA--AUUAUGCGACGUCA--CGGAAACGGCAUUUUAUUGAAUGU----CAACUGCAACUCGAGU-CUGCGAUUAUUGACCAC------- --------...(((.(((((((((((((..--....((((......--.(....)(((((((....))))))----)...))))......))-)))))).))))).))).------- ( -31.00, z-score = -3.55, R) >droYak2.chr3L 3481868 93 + 24197627 --------GAUUGGACAAUAUUGCAGGCGA--AUUAUGCGACGUCA--CGGAAACGGCAUUUUAUUGAAUGU----CAACUGCAACUCGAGU-CUGCGAUUAUUGACCAC------- --------...(((.(((((((((((((..--....((((......--.(....)(((((((....))))))----)...))))......))-)))))).))))).))).------- ( -30.60, z-score = -3.88, R) >droEre2.scaffold_4784 19604104 93 - 25762168 --------AAUUGGACAAUAUUGCAGGCGA--AUUAUGCGACGUCA--CGGAAACGGCAUUUUAUUGAAUGU----CAACUGCAACUCGAGU-CUGCGAUUAUUGACCAC------- --------...(((.(((((((((((((..--....((((......--.(....)(((((((....))))))----)...))))......))-)))))).))))).))).------- ( -30.60, z-score = -3.95, R) >dp4.chrXR_group6 1487823 97 + 13314419 AUUUGAGCAACUGCAAAAUAGCAGAGA-GA--AUUAUGCGACGUCA--CGGAAACGGAAUUUUAUUGAA-------AAGCUGCAAUUCUGCU-UCAUGUUCUCUGUUUUC------- ....((((..((((......))))(((-((--((...(((......--((....))(((((.((((...-------.)).)).)))))))).-....)))))))))))..------- ( -21.80, z-score = -0.39, R) >droPer1.super_20 123918 97 + 1872136 AUUUGAGCAACUGCAAAAUAGUAGAGA-GA--AUUAUGCGACGUCA--CGGAAACGGAAUUUUAUUGAA-------AAGCUGCAAUUCUGCU-UCAUGUUCUCUGUUUUC------- ....((((..((((......))))(((-((--((...(((......--((....))(((((.((((...-------.)).)).)))))))).-....)))))))))))..------- ( -19.80, z-score = 0.01, R) >droVir3.scaffold_13049 2352515 108 + 25233164 ---------AUUAUCUCUGAUUGCAAAGAAUUAUUAUGCGACGUCACGCGACAACGGUAUUUCAUAGCAGGUUUUGCUGUUGUCUUUUUAUUGUUGUCGUUUUUGUUUUUUUGCUGC ---------.............((((((((.......(((......)))((((((((((....(((((((......))))))).....))))))))))........))))))))... ( -28.70, z-score = -2.79, R) >consensus ________GAUUGGACAAUAUUGCAGGCGA__AUUAUGCGACGUCA__CGGAAACGGCAUUUUAUUGAAUGU____CAACUGCAACUCGAGU_CUGCGAUUAUUGACCAC_______ ...................(((((((.(((......((((....)...((....)).........................)))..)))....)))))))................. ( -9.26 = -8.46 + -0.79)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:39 2011