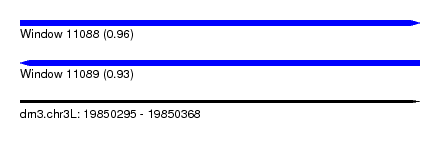
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,850,295 – 19,850,368 |
| Length | 73 |
| Max. P | 0.960556 |

| Location | 19,850,295 – 19,850,368 |
|---|---|
| Length | 73 |
| Sequences | 3 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 57.35 |
| Shannon entropy | 0.57744 |
| G+C content | 0.42377 |
| Mean single sequence MFE | -15.87 |
| Consensus MFE | -12.03 |
| Energy contribution | -11.27 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
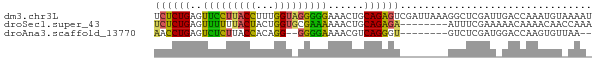
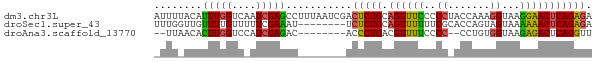
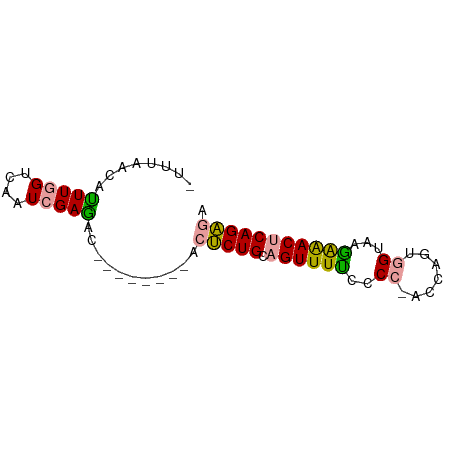
>dm3.chr3L 19850295 73 + 24543557 UCUCUGAGUUCCUUACCUUUGGUAGGGGGAAACUGCAGAGUCGAUUAAAGGCUCGAUUGACCAAAUGUAAAAU ............((((.((((((.(.((....)).).(((((.......))))).....)))))).))))... ( -20.60, z-score = -1.70, R) >droSec1.super_43 270810 65 - 327307 UCUCUGAGUUUUUUACUACUGGUGCGAAAAAACUGCAGAGA--------AUUUCGAAAAACAAAACAACCAAA ((((((((((((((..((....))..)))))))).))))))--------........................ ( -13.30, z-score = -2.39, R) >droAna3.scaffold_13770 628024 61 - 865660 AACCUGAGUCUCUUACCACAGG--GGGGAAAACGUCAGGGU--------GUCUCGAUGGACCAAGUGUUAA-- ..(((((((.(((..((...))--..)))..)).)))))(.--------((((....))))).........-- ( -13.70, z-score = 1.17, R) >consensus UCUCUGAGUUUCUUACCACUGGU_GGGGAAAACUGCAGAGU________GUCUCGAUAGACCAAAUGUAAAA_ .(((((..(((((((((...)))))))))......)))))................................. (-12.03 = -11.27 + -0.77)
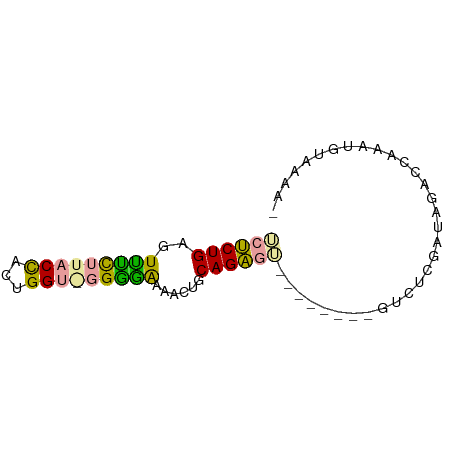
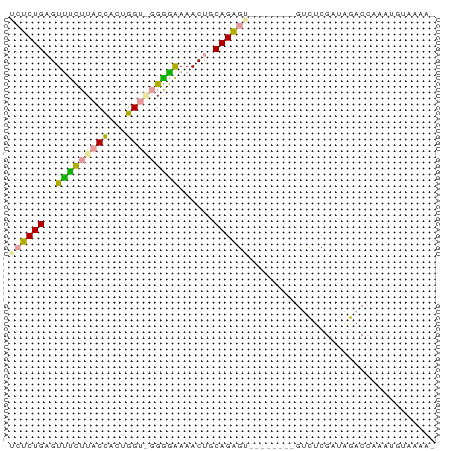
| Location | 19,850,295 – 19,850,368 |
|---|---|
| Length | 73 |
| Sequences | 3 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 57.35 |
| Shannon entropy | 0.57744 |
| G+C content | 0.42377 |
| Mean single sequence MFE | -15.30 |
| Consensus MFE | -8.64 |
| Energy contribution | -8.20 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.929486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19850295 73 - 24543557 AUUUUACAUUUGGUCAAUCGAGCCUUUAAUCGACUCUGCAGUUUCCCCCUACCAAAGGUAAGGAACUCAGAGA .................((((........))))(((((.((((((..(((.....)))...))))))))))). ( -17.90, z-score = -1.93, R) >droSec1.super_43 270810 65 + 327307 UUUGGUUGUUUUGUUUUUCGAAAU--------UCUCUGCAGUUUUUUCGCACCAGUAGUAAAAAACUCAGAGA .......((((((.....))))))--------((((((.((((((((.((.......)))))))))))))))) ( -15.10, z-score = -2.19, R) >droAna3.scaffold_13770 628024 61 + 865660 --UUAACACUUGGUCCAUCGAGAC--------ACCCUGACGUUUUCCCC--CCUGUGGUAAGAGACUCAGGUU --......(((((....)))))..--------..(((((.(((((...(--(....))...)))))))))).. ( -12.90, z-score = 0.23, R) >consensus _UUUAACAUUUGGUCAAUCGAGAC________ACUCUGCAGUUUUCCCC_ACCAGUGGUAAGAAACUCAGAGA ........(((((....)))))...........(((((.((((((..((.......))...))))))))))). ( -8.64 = -8.20 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:37 2011