
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,821,582 – 19,821,683 |
| Length | 101 |
| Max. P | 0.792129 |

| Location | 19,821,582 – 19,821,683 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.32 |
| Shannon entropy | 0.66437 |
| G+C content | 0.52442 |
| Mean single sequence MFE | -33.96 |
| Consensus MFE | -13.45 |
| Energy contribution | -12.90 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.70 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
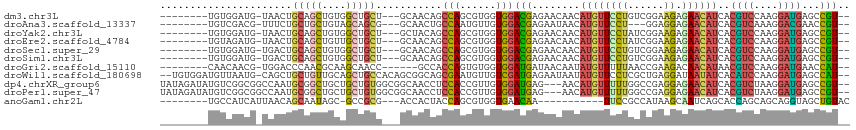
>dm3.chr3L 19821582 101 + 24543557 --------UGUGGAUG-UAACUGCAGCUGUGGCUGCU---GCAACAGCCAGCGUGGUGGACGAGAACAACAUGUUCCUGUCGGAAGAGAACAUCACGUCCAAGGAUGAGCCGU-- --------..((((((-(.((..(.(((..(((((..---....)))))))))..)).((((.((((.....)))).)))).............))))))).((.....))..-- ( -32.90, z-score = -0.40, R) >droAna3.scaffold_13337 3858962 98 - 23293914 --------UGUCGACG-UUUCUGCUGCUGUAGCAGCG---GCAACUGCCAAUGUUGUGGACGAGAAUAACAUGUUCCU---GGAGGAGAACAUCACGUCAAAGGACGAACCGU-- --------.(((.(((-.....(((((....)))))(---((....))).....))).))).........(((((((.---....).))))))..((((....))))......-- ( -29.00, z-score = -0.48, R) >droYak2.chr3L 3448675 101 - 24197627 --------UGUGGAUG-UAACUGCAGCUGUGGCUGCU---GCUACAGCCAGCGUGGUGGACGAGAACAACAUGUUCCUAUCGGAAGAGAACAUCACGUCCAAGGAUGAGCCGU-- --------..(((((.-.....((.((((((((....---))))))))..))((((((..(..((((.....))))...((....)))..))))))))))).((.....))..-- ( -36.60, z-score = -1.83, R) >droEre2.scaffold_4784 19573310 101 + 25762168 --------UGUAGAUG-UAACUGCAGCUGUUGCUGCU---GCAACAGCCAGCGUGGUGGACGAGAACAACAUGUUCCUAUCGGAAGAGAACAUCACGUCCAAGGAUGAGCCGU-- --------(((((...-...)))))((((((((....---))))))))..((((((((..(..((((.....))))...((....)))..))))))))....((.....))..-- ( -35.40, z-score = -2.12, R) >droSec1.super_29 475296 101 + 700605 --------UGUGGAUG-UGACUGCAGCUGUGGCUGCU---GCAACAGCCAGCGUGGUGGACGAGAACAACAUGUUCCUGUCGGAAGAGAACAUCACGUCCAAGGAUGAGCCGU-- --------..((((((-((((..(.(((..(((((..---....)))))))))..)).((((.((((.....)))).))))............)))))))).((.....))..-- ( -35.80, z-score = -0.98, R) >droSim1.chr3L 19106245 101 + 22553184 --------UGUGGAUG-UGACUGCAGCUGUGGCUGCU---GCAACAGCCAGCGUGGUGGACGAGAACAACAUGUUCCUGUCGGAAGAGAACAUCACGUCCAAGGAUGAGCCGU-- --------..((((((-((((..(.(((..(((((..---....)))))))))..)).((((.((((.....)))).))))............)))))))).((.....))..-- ( -35.80, z-score = -0.98, R) >droGri2.scaffold_15110 6380910 98 - 24565398 --------CAACAACG-UGGACCCAACGCAAGCAACC------GCCACCAGUGUGGUGGAUGAUAACAAUAUGUUUUUAACCGAAGACAACAUAACGUCCAAGGAUGAACCAU-- --------........-(((((....(....)((.((------(((((....))))))).)).........(((((((....))))))).......))))).((.....))..-- ( -23.80, z-score = -1.62, R) >droWil1.scaffold_180698 8653278 110 - 11422946 --UGUGGAUGUUAAUG-CAGCUGCUGUUGCAGCUGCCACAGCGGCAGCGAAUGUUGUCGAUGAGAAUAAUAUGUUCCUCGCUGAGGAUAAUAUCACAUCCAAGGAUGAGCCAU-- --.((((((((((..(-(((((((....))))))))..((((((((((....))))))...((((((.....))).)))))))....))))))).((((....))))...)))-- ( -41.60, z-score = -2.94, R) >dp4.chrXR_group6 5936603 110 + 13314419 UAUAGAUAUGUCGGCGGCCAAUGCGGCUGCUGCUGUGGCGGCAACCUCCACCGUUGUGGAUGAG---AACAUGUUUUUGGCCGAGGAGAACAUCACGUCUAAGGAUGAGCCGU-- ...........((((((((((.((((.((((((....)))))).))(((((....)))))....---.....))..))))))........((((.(......))))).)))).-- ( -40.20, z-score = -1.42, R) >droPer1.super_47 436663 110 + 592741 UAUAGAUAUGUCGGCGGCCAAUGCGGCUGCUGCUGUGGCGGCAACCUCCACCGUUGUGGAUGAG---AACAUGUUUUUGGCCGAGGAGAACAUCACGUCUAAGGAUGAGCCGU-- ...........((((((((((.((((.((((((....)))))).))(((((....)))))....---.....))..))))))........((((.(......))))).)))).-- ( -40.20, z-score = -1.42, R) >anoGam1.chr2L 5271069 92 + 48795086 --------UGCCAUCAUUAACAGCAAUAGC-GCCGCG---ACCACUACCAGCGUGGUGAACAA-----------UUCCGCCAUAAGCAAUCAGCACCAGCAGCAGGUAGCUGUAC --------...........(((((....((-...)).---.....((((.((((((((.....-----------...))))))..((.....)).......)).))))))))).. ( -22.30, z-score = -0.54, R) >consensus ________UGUCGAUG_UAACUGCAGCUGUGGCUGCU___GCAACAGCCAGCGUGGUGGACGAGAACAACAUGUUCCUGUCGGAAGAGAACAUCACGUCCAAGGAUGAGCCGU__ ......................(((((....)))))...........(((......)))(((........((((((((......)).))))))..((((....))))...))).. (-13.45 = -12.90 + -0.55)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:35 2011