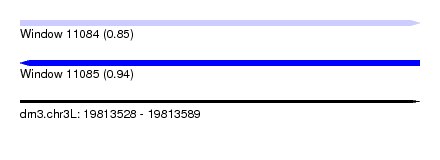
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,813,528 – 19,813,589 |
| Length | 61 |
| Max. P | 0.938187 |

| Location | 19,813,528 – 19,813,589 |
|---|---|
| Length | 61 |
| Sequences | 3 |
| Columns | 69 |
| Reading direction | forward |
| Mean pairwise identity | 61.08 |
| Shannon entropy | 0.53471 |
| G+C content | 0.44805 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -4.59 |
| Energy contribution | -6.60 |
| Covariance contribution | 2.01 |
| Combinations/Pair | 1.37 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
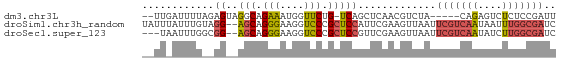
>dm3.chr3L 19813528 61 + 24543557 --UUGAUUUUAGAGUAGGCAGAAAUGGUUCUG-UCAGCUCAACGUCUA-----CAGAGUCUCUCCGAUU --..((((((.((((.(((((((....)))))-)).))))...(....-----)))))))......... ( -15.70, z-score = -1.22, R) >droSim1.chr3h_random 100551 67 - 1452968 UAUUUAUUUGUAGG--AGCAGGGAAGGUCCCGCUCCAUUCGAAGUUAAUUCGUCAAUAAUUUGGCGAUC ......((((..((--(((.(((.....))))))))...))))......(((((((....))))))).. ( -23.10, z-score = -4.17, R) >droSec1.super_123 67620 64 + 76498 ---UAAUUUGGCGG--AGCAGGGAAGGUCCCGCUCCGUUCGAAGUUAAUUCGUCAAUAUCUUGGCGAUC ---(((((((((((--(((.(((.....)))))))))))..))))))..(((((((....))))))).. ( -26.70, z-score = -4.10, R) >consensus __UUAAUUUGAAGG__AGCAGGGAAGGUCCCGCUCCGUUCGAAGUUAAUUCGUCAAUAUCUUGGCGAUC ......((((((....(((.(((....))).)))...))))))......(((((((....))))))).. ( -4.59 = -6.60 + 2.01)


| Location | 19,813,528 – 19,813,589 |
|---|---|
| Length | 61 |
| Sequences | 3 |
| Columns | 69 |
| Reading direction | reverse |
| Mean pairwise identity | 61.08 |
| Shannon entropy | 0.53471 |
| G+C content | 0.44805 |
| Mean single sequence MFE | -16.67 |
| Consensus MFE | -5.86 |
| Energy contribution | -5.87 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
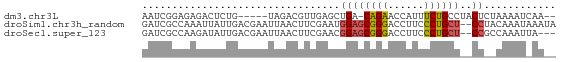
>dm3.chr3L 19813528 61 - 24543557 AAUCGGAGAGACUCUG-----UAGACGUUGAGCUGA-CAGAACCAUUUCUGCCUACUCUAAAAUCAA-- ...(((((...)))))-----..((..(((((..(.-(((((....))))).)..))).))..))..-- ( -11.10, z-score = -0.24, R) >droSim1.chr3h_random 100551 67 + 1452968 GAUCGCCAAAUUAUUGACGAAUUAACUUCGAAUGGAGCGGGACCUUCCCUGCU--CCUACAAAUAAAUA ..(((.(((....))).))).............((((((((......))))))--))............ ( -18.00, z-score = -3.84, R) >droSec1.super_123 67620 64 - 76498 GAUCGCCAAGAUAUUGACGAAUUAACUUCGAACGGAGCGGGACCUUCCCUGCU--CCGCCAAAUUA--- ..(((.(((....))).)))............(((((((((......))))))--)))........--- ( -20.90, z-score = -3.61, R) >consensus GAUCGCCAAGAUAUUGACGAAUUAACUUCGAACGGAGCGGGACCUUCCCUGCU__CCCACAAAUAAA__ ...................................((((((......))))))................ ( -5.86 = -5.87 + 0.00)
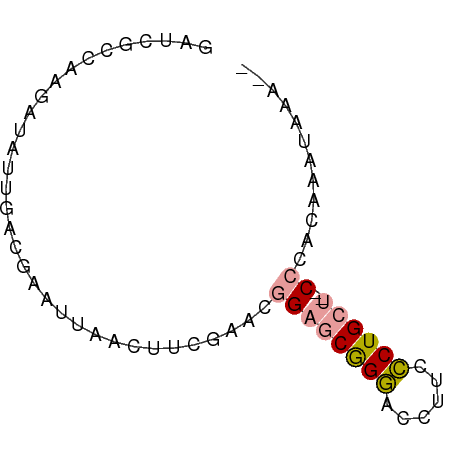

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:34 2011