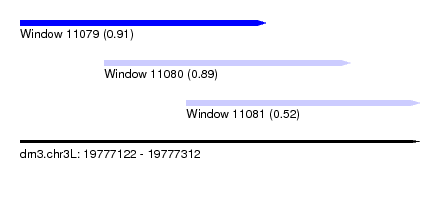
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,777,122 – 19,777,312 |
| Length | 190 |
| Max. P | 0.905363 |

| Location | 19,777,122 – 19,777,239 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.29 |
| Shannon entropy | 0.30094 |
| G+C content | 0.43320 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.68 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19777122 117 + 24543557 UGAUUUUGCUUUUUAUCGCUCCUGGAUUGGUUCUUUCGUCUGCCUUCAAUUCAACUUCUCGCCGAAACUUUUGGCCGCGGAAAA-GUUGAAUAAACUUGGCCGAAAGGCCAGAAGUGA ...............(((((.((((......(((((((...(((....(((((((((.(((((((.....)))))....)).))-)))))))......)))))))))))))).))))) ( -33.70, z-score = -1.47, R) >droSim1.chr3L 19068581 108 + 22553184 UGACCUUUCUUUUUAUCGCUUCUGGAUUGGUUCUUUC----------AGUUCAAAUUCUCGCCGAAACUUUUGGCCGCGAAAAAAGUUUAAUAAACUUGACCGAAAGGCCAGAAGUGA ...............((((((((((.((((((.....----------........(((.((((((.....)))).)).)))..(((((.....)))))))))))....)))))))))) ( -28.70, z-score = -1.77, R) >droSec1.super_29 438197 108 + 700605 UGAUUUUGCUUUUUAUCGCUUCUGGAUUGGUUUUUUC----------AGUUCAACUUCUCGCCGAAACUUUUGGCCGCGAAAAAAGUUUAAUAAACUUGGCCGAAAGGCCAGAAGUGA ..(((..(((((((.((((...((((((((.....))----------)))))).......(((((.....))))).)))))))))))..)))....((((((....))))))...... ( -36.20, z-score = -3.49, R) >droYak2.chr3L 3411466 114 - 24197627 UGAACUUGCUCUUUAACGCUUCUGGGUUGGCUUCAUC---UGUCUUCAGUUAAACUUAUCGCCGAAACUUUUGGGCGCGGAAAA-GUUAAGUAAACUUGGCCGAAAUGCCAGGAGUGU ...............((((((((((.((((((...((---(((....((.....))....(((.((....)).)))))))).((-(((.....)))))))))))....)))))))))) ( -31.50, z-score = -0.86, R) >droEre2.scaffold_4784 19535654 113 + 25762168 UGAUUCUGCCCUUUUUCGCUCCUGGGUUGGCUUCAUC---UCUCUUCGGUUCAACUUCUCGCCGAA-CUUUUGGCCGCGGAAAA-GUUUAAUAAACUUGGCCGAAAGGCCAGAAUUGA .((((((...(((((((((.((.((((((((......---...(....)...........))).))-)))..))..))))))))-)............((((....)))))))))).. ( -35.15, z-score = -1.90, R) >consensus UGAUUUUGCUUUUUAUCGCUUCUGGAUUGGUUUUUUC___U__CUUCAGUUCAACUUCUCGCCGAAACUUUUGGCCGCGGAAAA_GUUUAAUAAACUUGGCCGAAAGGCCAGAAGUGA ...............((((((((((.(((((.............................)))))..((((((((((.(................).)))))))))).)))))))))) (-25.12 = -25.68 + 0.56)

| Location | 19,777,162 – 19,777,279 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 64.73 |
| Shannon entropy | 0.69296 |
| G+C content | 0.42143 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -10.32 |
| Energy contribution | -11.27 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19777162 117 + 24543557 UGCCUUCAAUUCAACUUCUCGCCGAAACUUUUGGCCGCGGAAAA-GUUGAAUAAACUUGGCCGAAAGGCCAGAAGUGAGCUUAAUUGACGAUGGGGUAAUUAUUACUUCUGCCAAUGC .((((((.(((((((((.(((((((.....)))))....)).))-))))))).....(((((....)))))))))...))...((((.((..((((((.....)))))))).)))).. ( -35.20, z-score = -2.28, R) >droSim1.chr3L 19068615 114 + 22553184 ----UUCAGUUCAAAUUCUCGCCGAAACUUUUGGCCGCGAAAAAAGUUUAAUAAACUUGACCGAAAGGCCAGAAGUGAGCUUAAUUGGCGAUGAGGUAUUUAUUACUUCUGCCCAAGC ----...(((((...(((.....)))((((((((((.((....(((((.....)))))...))...))))))))))))))).....((((..((((((.....))))))))))..... ( -34.10, z-score = -3.00, R) >droSec1.super_29 438231 114 + 700605 ----UUCAGUUCAACUUCUCGCCGAAACUUUUGGCCGCGAAAAAAGUUUAAUAAACUUGGCCGAAAGGCCAGAAGUGAGCUUAAUUGACGUUGGGGUAUUUAUUACUUCUGCCCAAGC ----...((((((.(((((.(((((.....)))))........(((((.....)))))((((....)))))))))))))))........(((.(((((...........))))).))) ( -34.30, z-score = -2.26, R) >droYak2.chr3L 3411503 117 - 24197627 UGUCUUCAGUUAAACUUAUCGCCGAAACUUUUGGGCGCGGAAAA-GUUAAGUAAACUUGGCCGAAAUGCCAGGAGUGUGCUUUAUUGUCGAUUGGGUAUUUAUGACUUCUUCACAAAC (((....(((((....(((((((.((....)).))).(((...(-((.(((((..((((((......))))))....))))).))).)))....))))....))))).....)))... ( -24.80, z-score = 0.46, R) >droEre2.scaffold_4784 19535691 96 + 25762168 UCUCUUCGGUUCAACUUCUCGCCGAA-CUUUUGGCCGCGGAAAA-GUUUAAUAAACUUGGCCGAAAGGCCAGAAUUGAGCUUUAUUGUCGAUGGAGUA-------------------- .(((((((............(((((.-...))))).((((.(((-(((((((....((((((....)))))).)))))))))).))))))).))))..-------------------- ( -32.40, z-score = -2.67, R) >droAna3.scaffold_13337 6136683 99 - 23293914 ----------------UCCCGCAUAGUAUGCCACAAACAAGCAAACAAAGGAACAAUUAGACAUAAUUCCAUCAGGAAAC---AUUGGCACUUGAAGACCUCUGAUUGUUUUGUUCAU ----------------.........((.(((.........))).))...((((((((((((.....((((....))))..---...(((.......).))))))))))))))...... ( -17.10, z-score = -0.48, R) >consensus ____UUCAGUUCAACUUCUCGCCGAAACUUUUGGCCGCGAAAAA_GUUUAAUAAACUUGGCCGAAAGGCCAGAAGUGAGCUUAAUUGACGAUGGGGUAUUUAUUACUUCUGCCCAAGC ....................((....((((((((((.((......................))...))))))))))..))...................................... (-10.32 = -11.27 + 0.95)


| Location | 19,777,201 – 19,777,312 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.35 |
| Shannon entropy | 0.71517 |
| G+C content | 0.35770 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -6.79 |
| Energy contribution | -7.35 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.524689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19777201 111 + 24543557 GAAAAGUUGAAUAAAC--UUGGCCGAAAGGCCAGAAGUGAGCUUAAUUGACGAUGGGGUAAUUAUUACUUCUGCCAAUGCAUUUUGGUAUGAAAAUAAGAGCAUGCAUAUUGA------- ...(((((.....)))--))((((....))))((((((((...((((((.(......)))))))))))))))....((((((.((..(((....)))..)).)))))).....------- ( -30.10, z-score = -2.94, R) >droSim1.chr3L 19068651 111 + 22553184 AAAAAGUUUAAUAAAC--UUGACCGAAAGGCCAGAAGUGAGCUUAAUUGGCGAUGAGGUAUUUAUUACUUCUGCCCAAGCAUUUUGCUAUGAAAAUUAGUGCAUGCAUAUUGA------- ...(((((((......--(((.((....)).)))...)))))))....((((..((((((.....))))))))))...((((...((((.......))))..)))).......------- ( -25.50, z-score = -1.37, R) >droSec1.super_29 438267 110 + 700605 AAAAAGUUUAAUAAAC--UUGGCCGAAAGGCCAGAAGUGAGCUUAAUUGACGUUGGGGUAUUUAUUACUUCUGCCCAAGCAUUUUGCUAUGAAAAUUAGUGCAUGC-UAUUGA------- ...(((((((......--((((((....))))))...))))))).......((.((((((.....)))))).))...(((((...((((.......))))..))))-).....------- ( -29.20, z-score = -1.91, R) >droYak2.chr3L 3411542 118 - 24197627 GAAAAGUUAAGUAAAC--UUGGCCGAAAUGCCAGGAGUGUGCUUUAUUGUCGAUUGGGUAUUUAUGACUUCUUCACAAACAUUUUAGUGUGAAAAUAAGAGAUUAAAGAUUGAAUAAAUC ....(((.(((((..(--(((((......))))))....))))).))).((((((....((((........((((((..........))))))......))))....))))))....... ( -19.94, z-score = 0.05, R) >droEre2.scaffold_4784 19535729 96 + 25762168 GAAAAGUUUAAUAAAC--UUGGCCGAAAGGCCAGAAUUGAGCUUUAUUGUCGAUGGAGUAUUUA----------------------GUAUCAAAAUGGGAGCAUGAAGAUUGAAUAAAUC ..((((((((((....--((((((....)))))).))))))))))......(((....((((((----------------------((......(((....)))....)))))))).))) ( -26.10, z-score = -4.08, R) >droAna3.scaffold_13337 6136705 101 - 23293914 AAGCAAACAAAGGAACAAUUAGACAUAAUUCCAUCAGGAAAC---AUUGGCACUUGAAGACCUCUGAUUGUUUUGUUCAUCUUGUCACAUGCCAUGUCCCUCAG---------------- ..........(((.(((...........((((....))))..---..(((((..((((((....(((.........))))))..)))..)))))))).)))...---------------- ( -16.40, z-score = 0.83, R) >consensus AAAAAGUUUAAUAAAC__UUGGCCGAAAGGCCAGAAGUGAGCUUAAUUGACGAUGGGGUAUUUAUUACUUCUGCCCAAGCAUUUUGGUAUGAAAAUUAGAGCAUGCAUAUUGA_______ ..................((((((....))))))...................................................................................... ( -6.79 = -7.35 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:30 2011