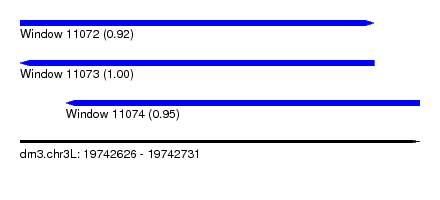
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,742,626 – 19,742,731 |
| Length | 105 |
| Max. P | 0.999006 |

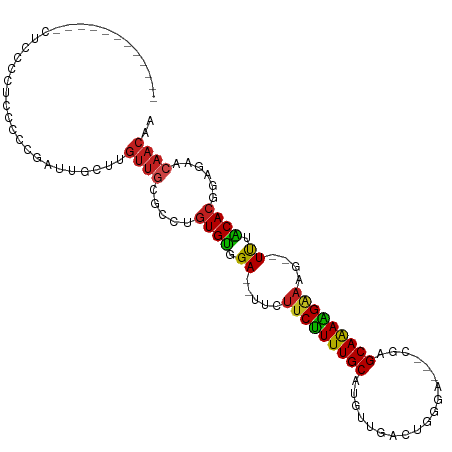
| Location | 19,742,626 – 19,742,719 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 66.48 |
| Shannon entropy | 0.66583 |
| G+C content | 0.47544 |
| Mean single sequence MFE | -26.29 |
| Consensus MFE | -15.33 |
| Energy contribution | -15.28 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
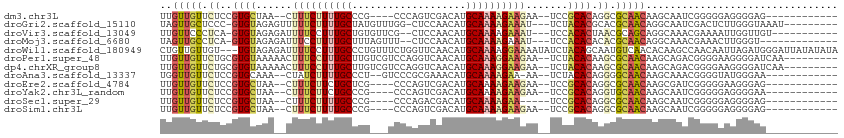
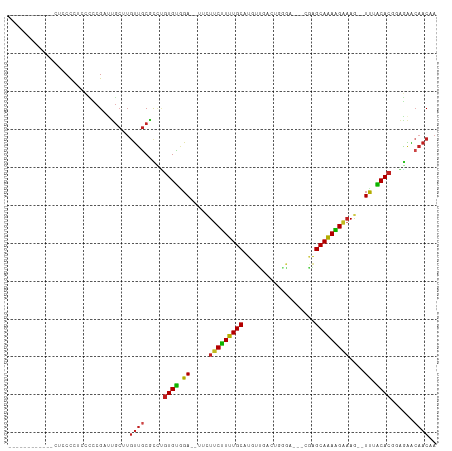
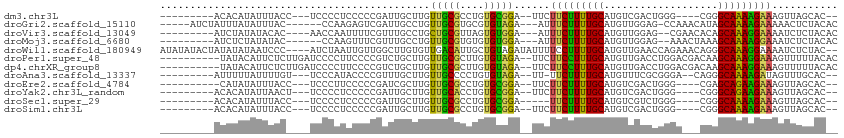
>dm3.chr3L 19742626 93 + 24543557 UUGUUGUUCUCCGUGCUAA--CUUUCUUUUGCCCG----CCCAGUCGACAUGCAAAAGAAGAA--UCCGCACAGGCGCAACAAGCAAUCGGGGGAGGGGAG------------ .(.((.(((((((((((..--.((((((((((.((----......))....))))))))))..--..(((....))).....))))..))))))).)).).------------ ( -25.20, z-score = 0.48, R) >droGri2.scaffold_15110 15532712 99 + 24565398 UAGUUGCUCCC-GUGUAGAGUUUUUCUUUUGCUAUGUUUGG-CUCCAACAUGCAAAAGAAAU---UCUACACGCACGCAACAGGCAAUCGACUCUUGGGUAAAU--------- ..(((((...(-((((((((((..((((((((.(((((.(.-...)))))))))))))))))---))))))))...))))).........(((....)))....--------- ( -36.20, z-score = -5.01, R) >droVir3.scaffold_13049 11529058 96 - 25233164 UUGUUCCCUCA-GUGUAGAGAUUUUCCUUUGCUGUGUUCG--CUCCAACAUGCAAAAGAAAU---UCCACACUAACGCAGCAGGCAAACGAAAAUUGGUUGU----------- ..........(-((((.(.((.((((.(((((.(((((..--....)))))))))).)))).---))))))))......((((.(((.......))).))))----------- ( -20.80, z-score = -0.39, R) >droMoj3.scaffold_6680 13642635 94 + 24764193 UAGUUGCCUCA-GUGUAGAGAUUUCCUUUUGCUUUAGUUU--CUCCAACAUGCAAAAGAAAU---UCCACACACACGCAACAGGCAAACGAAACUUGGGU------------- ..(((((....-((((.(..(((((.((((((....(((.--....)))..)))))))))))---..)))))....)))))...................------------- ( -21.10, z-score = -1.15, R) >droWil1.scaffold_180949 4853085 110 + 6375548 CUGUUGUUGU---UGUAGAGAUUUUCCUUUGCCCUGUUUCUGGUUCAACAUGCAAAAGGAAAAUAUCUACAGCAAUGUCAACACAAGCCAACAAUUAGAUGGGAUUAUAUAUA .(((((((((---((((((.((((((((((((..((((........)))).)).)))))))))).))))))))))...)))))....(((.(.....).)))........... ( -31.70, z-score = -3.12, R) >droPer1.super_48 566603 102 + 581423 UUGUUGUUCUGCGUGUAAAAACUUUCCUUUGCUUGUCGUCCAGGUCAACAUGCAAAGGAAGAA--UCUACACAAGCGCAACAAGCAGACGGGGAAGGGGAUCAA--------- (((((((.((..(((((.....((((((((((.(((.(.......).))).))))))))))..--..))))).)).))))))).....................--------- ( -28.40, z-score = -1.07, R) >dp4.chrXR_group8 7785195 102 + 9212921 UUGUUGUUCUGCGUGUAAAAACUUUCCUUUGCUUGUCGUCCAGGUCAACAUGCAAAGGAAGAA--UCUACACAAGCGCAACAAGCAGACGGGGAAGGGGAUCAA--------- (((((((.((..(((((.....((((((((((.(((.(.......).))).))))))))))..--..))))).)).))))))).....................--------- ( -28.40, z-score = -1.07, R) >droAna3.scaffold_13337 6109106 94 - 23293914 UGGUUGUUCUCCGUGCAAA--CUAUCUUUUGCCCU--GUCCCGCGAAACAUGCAAAAGAA-AA--UCUACACAGGGGCAACAAGCAAACGGGGUAUGGGAA------------ (((((((.......)).))--)))(((..((((((--((((((((.....)))...((..-..--.)).....)))((.....))..))))))))..))).------------ ( -23.90, z-score = -0.00, R) >droEre2.scaffold_4784 19504143 93 + 25762168 UUGUUGUUCUCCGUGCUAA--CUUUCUUCUGCUCG----CCCAGUCGACAUGCAAAAGAAGAA--UCCGCACAGGCGCAACAAGCGAUCGGGGGAAGGGAG------------ ...((.((((((((((...--..((((((((((((----......)))...))...)))))))--...)))).(((((.....))).)).)))))).))..------------ ( -25.60, z-score = 0.33, R) >droYak2.chr3L_random 3342874 93 + 4797643 UUGUUGUUCUCCGUGCUAA--CUUUCUUCUGCCCG----CCCAGUCGACAUGCAAAAGAAGAA--UCCGCACAGGUGCAACAAGCAAUCGGGGGAGGGGAA------------ .(.((.(((((((((((..--..(((((((....(----(...........))...)))))))--...(((....)))....))))..))))))).)).).------------ ( -23.90, z-score = 0.90, R) >droSec1.super_29 409242 90 + 700605 UUGUUGUUCUCCGUGCUAA--CUUUCUUUUGCCCG----CCCAGACGACAUGCAAAAGAA-----UCCGCACAGGCGCAACAAGCAAUCGGGGGAGGGGAG------------ .(.((.(((((((((((..--..(((((((((.((----......))....)))))))))-----..(((....))).....))))..))))))).)).).------------ ( -25.10, z-score = 0.24, R) >droSim1.chr3L 19037289 93 + 22553184 UUGUUGUUCUCCGUGCUAA--CUUUCUUUUGCCCG----CCCAGUCGACAUGCAAAAGAAGAA--UCCGCACAGGCGCAACAAGCAAUCGGGGGAGGGGAG------------ .(.((.(((((((((((..--.((((((((((.((----......))....))))))))))..--..(((....))).....))))..))))))).)).).------------ ( -25.20, z-score = 0.48, R) >consensus UUGUUGUUCUCCGUGCAAA__CUUUCUUUUGCCCG___UCCCAGUCAACAUGCAAAAGAAGAA__UCCACACAGGCGCAACAAGCAAUCGGGGGAGGGGAG____________ ..(((((.((..((((......((((((((((...................)))))))))).......)))).)).)))))................................ (-15.33 = -15.28 + -0.05)
| Location | 19,742,626 – 19,742,719 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 66.48 |
| Shannon entropy | 0.66583 |
| G+C content | 0.47544 |
| Mean single sequence MFE | -31.54 |
| Consensus MFE | -14.46 |
| Energy contribution | -13.84 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
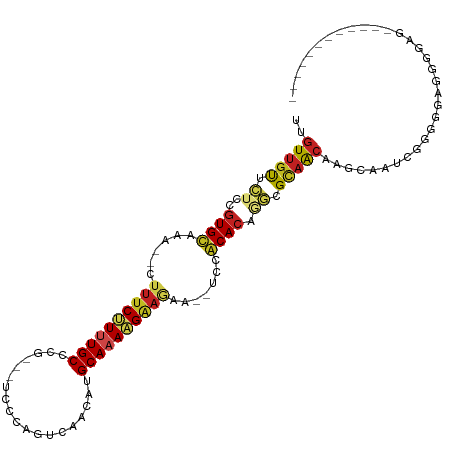
>dm3.chr3L 19742626 93 - 24543557 ------------CUCCCCUCCCCCGAUUGCUUGUUGCGCCUGUGCGGA--UUCUUCUUUUGCAUGUCGACUGGG----CGGGCAAAAGAAAG--UUAGCACGGAGAACAACAA ------------...................(((((..(((((((.((--((.(((((((((.((((.....))----)).)))))))))))--)).)))))).)..))))). ( -31.80, z-score = -1.70, R) >droGri2.scaffold_15110 15532712 99 - 24565398 ---------AUUUACCCAAGAGUCGAUUGCCUGUUGCGUGCGUGUAGA---AUUUCUUUUGCAUGUUGGAG-CCAAACAUAGCAAAAGAAAAACUCUACAC-GGGAGCAACUA ---------.......................(((((.(.((((((((---.((((((((((((((((...-.).))))).))))))))))...)))))))-).).))))).. ( -35.00, z-score = -4.26, R) >droVir3.scaffold_13049 11529058 96 + 25233164 -----------ACAACCAAUUUUCGUUUGCCUGCUGCGUUAGUGUGGA---AUUUCUUUUGCAUGUUGGAG--CGAACACAGCAAAGGAAAAUCUCUACAC-UGAGGGAACAA -----------....................((.(.(.((((((((((---.((((((((((.((((....--..))))..))))))))))...)))))))-))).).).)). ( -27.50, z-score = -1.61, R) >droMoj3.scaffold_6680 13642635 94 - 24764193 -------------ACCCAAGUUUCGUUUGCCUGUUGCGUGUGUGUGGA---AUUUCUUUUGCAUGUUGGAG--AAACUAAAGCAAAAGGAAAUCUCUACAC-UGAGGCAACUA -------------...................(((((.(..(((((((---.((((((((((...((((..--...)))).))))))))))...)))))))-..).))))).. ( -28.40, z-score = -2.33, R) >droWil1.scaffold_180949 4853085 110 - 6375548 UAUAUAUAAUCCCAUCUAAUUGUUGGCUUGUGUUGACAUUGCUGUAGAUAUUUUCCUUUUGCAUGUUGAACCAGAAACAGGGCAAAGGAAAAUCUCUACA---ACAACAACAG ....................(((..((....))..)))(((.((((((.((((((((((.((.((((........))))..)))))))))))).))))))---.)))...... ( -31.90, z-score = -3.80, R) >droPer1.super_48 566603 102 - 581423 ---------UUGAUCCCCUUCCCCGUCUGCUUGUUGCGCUUGUGUAGA--UUCUUCCUUUGCAUGUUGACCUGGACGACAAGCAAAGGAAAGUUUUUACACGCAGAACAACAA ---------......................(((((..((((((((((--..((((((((((.(((((.......))))).))))))).)))..)))))))).))..))))). ( -34.00, z-score = -4.25, R) >dp4.chrXR_group8 7785195 102 - 9212921 ---------UUGAUCCCCUUCCCCGUCUGCUUGUUGCGCUUGUGUAGA--UUCUUCCUUUGCAUGUUGACCUGGACGACAAGCAAAGGAAAGUUUUUACACGCAGAACAACAA ---------......................(((((..((((((((((--..((((((((((.(((((.......))))).))))))).)))..)))))))).))..))))). ( -34.00, z-score = -4.25, R) >droAna3.scaffold_13337 6109106 94 + 23293914 ------------UUCCCAUACCCCGUUUGCUUGUUGCCCCUGUGUAGA--UU-UUCUUUUGCAUGUUUCGCGGGAC--AGGGCAAAAGAUAG--UUUGCACGGAGAACAACCA ------------....................((((..((((((((((--((-.((((((((.(((((....))))--)..)))))))).))--))))))))).)..)))).. ( -32.00, z-score = -3.03, R) >droEre2.scaffold_4784 19504143 93 - 25762168 ------------CUCCCUUCCCCCGAUCGCUUGUUGCGCCUGUGCGGA--UUCUUCUUUUGCAUGUCGACUGGG----CGAGCAGAAGAAAG--UUAGCACGGAGAACAACAA ------------...................(((((..(((((((.((--((.(((((((((.((((.....))----)).)))))))))))--)).)))))).)..))))). ( -31.90, z-score = -1.89, R) >droYak2.chr3L_random 3342874 93 - 4797643 ------------UUCCCCUCCCCCGAUUGCUUGUUGCACCUGUGCGGA--UUCUUCUUUUGCAUGUCGACUGGG----CGGGCAGAAGAAAG--UUAGCACGGAGAACAACAA ------------...................(((((..(((((((.((--((.(((((((((.((((.....))----)).)))))))))))--)).)))))).)..))))). ( -30.60, z-score = -1.44, R) >droSec1.super_29 409242 90 - 700605 ------------CUCCCCUCCCCCGAUUGCUUGUUGCGCCUGUGCGGA-----UUCUUUUGCAUGUCGUCUGGG----CGGGCAAAAGAAAG--UUAGCACGGAGAACAACAA ------------...................(((((..(((((((...-----(((((((((.((((.....))----)).)))))))))..--...)))))).)..))))). ( -29.60, z-score = -1.07, R) >droSim1.chr3L 19037289 93 - 22553184 ------------CUCCCCUCCCCCGAUUGCUUGUUGCGCCUGUGCGGA--UUCUUCUUUUGCAUGUCGACUGGG----CGGGCAAAAGAAAG--UUAGCACGGAGAACAACAA ------------...................(((((..(((((((.((--((.(((((((((.((((.....))----)).)))))))))))--)).)))))).)..))))). ( -31.80, z-score = -1.70, R) >consensus ____________CUCCCCUCCCCCGAUUGCUUGUUGCGCCUGUGUGGA__UUCUUCUUUUGCAUGUUGACUGGGA___CGAGCAAAAGAAAG__UUUACACGGAGAACAACAA ................................((((.....((((.((.....(((((((((...................)))))))))....)).))))......)))).. (-14.46 = -13.84 + -0.62)

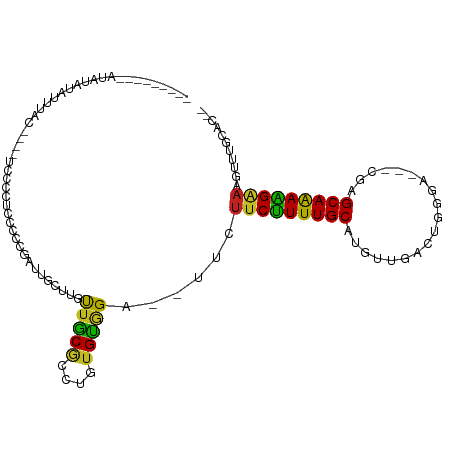
| Location | 19,742,638 – 19,742,731 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 64.93 |
| Shannon entropy | 0.68455 |
| G+C content | 0.44904 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -10.84 |
| Energy contribution | -9.89 |
| Covariance contribution | -0.95 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19742638 93 - 24543557 ---------ACACAUAUUUACC---UCCCCUCCCCCGAUUGCUUGUUGCGCCUGUGCGGA--UUCUUCUUUUGCAUGUCGACUGGG----CGGGCAAAAGAAAGUUAGCAC-- ---------.............---...............((.....))....((((.((--((.(((((((((.((((.....))----)).))))))))))))).))))-- ( -22.40, z-score = 0.03, R) >droGri2.scaffold_15110 15532723 98 - 24565398 -----AUCUAUUUAUAUUUAC------CCAAGAGUCGAUUGCCUGUUGCGUGCGUGUAGA---AUUUCUUUUGCAUGUUGGAG-CCAAACAUAGCAAAAGAAAAACUCUACAC -----.............(((------.(((.((........)).))).))).(((((((---.((((((((((((((((...-.).))))).))))))))))...))))))) ( -27.00, z-score = -3.37, R) >droVir3.scaffold_13049 11529069 95 + 25233164 ---------AUCUAUAUACAC----AACCAAUUUUCGUUUGCCUGCUGCGUUAGUGUGGA---AUUUCUUUUGCAUGUUGGAG--CGAACACAGCAAAGGAAAAUCUCUACAC ---------............----.....((((((.(((((.((((.((...(((..((---(....)))..)))..)).))--))......))))).))))))........ ( -20.70, z-score = -0.72, R) >droMoj3.scaffold_6680 13642646 93 - 24764193 ---------AUCUCUAUAUAC------CCAAGUUUCGUUUGCCUGUUGCGUGUGUGUGGA---AUUUCUUUUGCAUGUUGGAG--AAACUAAAGCAAAAGGAAAUCUCUACAC ---------...(((((((((------....((..((......))..))..)))))))))---.((((((((((...((((..--...)))).)))))))))).......... ( -21.30, z-score = -1.46, R) >droWil1.scaffold_180949 4853096 107 - 6375548 AUAUAUACUAUAUAUAAUCCC----AUCUAAUUGUUGGCUUGUGUUGACAUUGCUGUAGAUAUUUUCCUUUUGCAUGUUGAACCAGAAACAGGGCAAAGGAAAAUCUCUAC-- .((((((...)))))).....----.......(((..((....))..))).....(((((.((((((((((.((.((((........))))..)))))))))))).)))))-- ( -28.10, z-score = -2.90, R) >droPer1.super_48 566615 101 - 581423 ----------UAUACAUUCUCUUGAUCCCCUUCCCCGUCUGCUUGUUGCGCUUGUGUAGA--UUCUUCCUUUGCAUGUUGACCUGGACGACAAGCAAAGGAAAGUUUUUACAC ----------..............................((.....))....(((((((--..((((((((((.(((((.......))))).))))))).)))..))))))) ( -26.60, z-score = -2.95, R) >dp4.chrXR_group8 7785207 101 - 9212921 ----------UAUACAUUCUCUUGAUCCCCUUCCCCGUCUGCUUGUUGCGCUUGUGUAGA--UUCUUCCUUUGCAUGUUGACCUGGACGACAAGCAAAGGAAAGUUUUUACAC ----------..............................((.....))....(((((((--..((((((((((.(((((.......))))).))))))).)))..))))))) ( -26.60, z-score = -2.95, R) >droAna3.scaffold_13337 6109118 94 + 23293914 ---------AUUUUUAUUUUGU---UCCCAUACCCCGUUUGCUUGUUGCCCCUGUGUAGA--UU-UUCUUUUGCAUGUUUCGCGGGA--CAGGGCAAAAGAUAGUUUGCAC-- ---------.............---...............((.....))....(((((((--((-.((((((((.(((((....)))--))..)))))))).)))))))))-- ( -24.50, z-score = -2.12, R) >droEre2.scaffold_4784 19504155 92 - 25762168 ----------CAUAUAUUUACC---UCCCUUCCCCCGAUCGCUUGUUGCGCCUGUGCGGA--UUCUUCUUUUGCAUGUCGACUGGG----CGAGCAGAAGAAAGUUAGCAC-- ----------............---..............(((.....)))...((((.((--((.(((((((((.((((.....))----)).))))))))))))).))))-- ( -23.70, z-score = -0.80, R) >droYak2.chr3L_random 3342886 93 - 4797643 ---------ACACAUAUUAACU---UCCCCUCCCCCGAUUGCUUGUUGCACCUGUGCGGA--UUCUUCUUUUGCAUGUCGACUGGG----CGGGCAGAAGAAAGUUAGCAC-- ---------...........((---(((((.(((.(((((((.....)))...(((((((--.......)))))))))))...)))----.)))..))))...........-- ( -23.90, z-score = -0.56, R) >droSec1.super_29 409254 90 - 700605 ---------ACACAUAUUUACC---UCCCCUCCCCCGAUUGCUUGUUGCGCCUGUGCGGA-----UUCUUUUGCAUGUCGUCUGGG----CGGGCAAAAGAAAGUUAGCAC-- ---------.............---..............((((..(((((....))))).-----(((((((((.((((.....))----)).)))))))))....)))).-- ( -20.60, z-score = 0.70, R) >droSim1.chr3L 19037301 93 - 22553184 ---------ACACAUAUUUACC---UCCCCUCCCCCGAUUGCUUGUUGCGCCUGUGCGGA--UUCUUCUUUUGCAUGUCGACUGGG----CGGGCAAAAGAAAGUUAGCAC-- ---------.............---...............((.....))....((((.((--((.(((((((((.((((.....))----)).))))))))))))).))))-- ( -22.40, z-score = 0.03, R) >consensus _________AUAUAUAUUUAC____UCCCCUCCCCCGAUUGCUUGUUGCGCCUGUGUGGA__UUCUUCUUUUGCAUGUUGACUGGGA___CGAGCAAAAGAAAGUUUGCAC__ .............................................(((((....)))))......(((((((((...................)))))))))........... (-10.84 = -9.89 + -0.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:24 2011