| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,719,875 – 19,719,987 |
| Length | 112 |
| Max. P | 0.520322 |

| Location | 19,719,875 – 19,719,987 |
|---|---|
| Length | 112 |
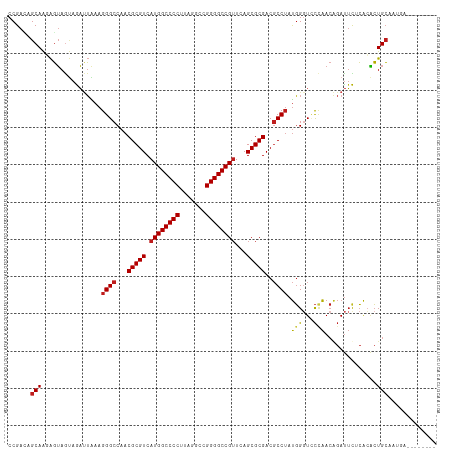
| Sequences | 13 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.79 |
| Shannon entropy | 0.34373 |
| G+C content | 0.57446 |
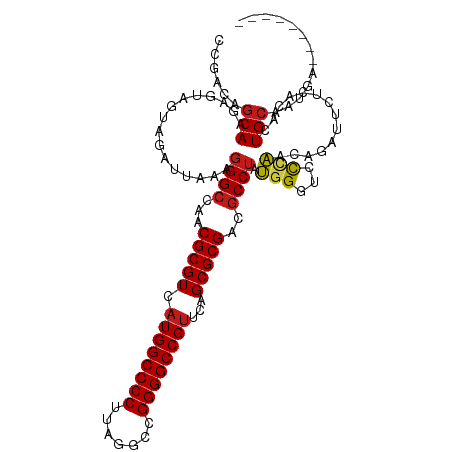
| Mean single sequence MFE | -38.11 |
| Consensus MFE | -29.39 |
| Energy contribution | -29.50 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.520322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19719875 112 + 24543557 CCGACAGCAAGAGUAGUAGAUUAAAGGGCCAACGCGUCAUGGCCCCUUAGGCCGGGGCCGUUCAGCGCGACGCCUAUGGGUCCCAACGGAUUCUCACACUGCAAUGAUGGGA--- (((.((......(((((........((((...(((((.((((((((.......))))))))...)))))..))))..((((((....))))))....)))))..)).)))..--- ( -40.90, z-score = -0.90, R) >droSim1.chr3L 19046070 112 + 22553184 CCGACAGCAAGAGUAGUAGAUUAAAGGGCCAACGCGUCAUGGCCCCUUAGGCCGGGGCCGUUCAGCGCGACGCCUAUGGGUCCCAACGGAUUCUCACACUGCAAUGAUGGGA--- (((.((......(((((........((((...(((((.((((((((.......))))))))...)))))..))))..((((((....))))))....)))))..)).)))..--- ( -40.90, z-score = -0.90, R) >droSec1.super_29 417534 112 + 700605 CCGACAGCAAGAGUAGUAGAUUAAAGGGCCAACGCGUCAUGGCCCCUUAGGCCGGGGCCGUUCAGCGCGACGCCUAUGGGUCCCAACGGAUUCUCACACUGCAAUGAAGGGA--- ((..((......(((((........((((...(((((.((((((((.......))))))))...)))))..))))..((((((....))))))....)))))..))..))..--- ( -40.80, z-score = -1.03, R) >droEre2.scaffold_4784 19512622 107 + 25762168 CCGACAGCAAGAGUAGUAGAUUAAAGGGCCAACGCGUCAUGGCCCCUUAGGCCGGGGCCGUUCAGCGCGACGCCUAUGGGUCCCAACGGAUUCUCACACUGCAAUGA-------- ............(((((........((((...(((((.((((((((.......))))))))...)))))..))))..((((((....))))))....))))).....-------- ( -39.70, z-score = -1.48, R) >droYak2.chr3L_random 3348928 107 + 4797643 CCGACAGCAAGAGUAGUAGAUUAAAGGGCCAACGCGUCAUGGCCCCUUAGGCCGGGGCCGUUCAGCGCGACGCCUAUGGGUUCCAACGGAUUCUCACACUGCAAUGA-------- ............(((((........((((...(((((.((((((((.......))))))))...)))))..)))).((((.(((...)))..)))).))))).....-------- ( -38.10, z-score = -1.18, R) >droAna3.scaffold_13337 6116565 111 - 23293914 CCGACAGCAAGAGUAAGAGACUAAAGGGCCAACGCGUCAUGGCCCCUUAGGCCGGGGCCGUUCAGCGCGACGCCUAUGGGUCCCAACAGAUUCUCACACUGCAACAAGAGA---- ......((((((((..(.((((...((((...(((((.((((((((.......))))))))...)))))..))))...)))).).....))))).....))).........---- ( -37.70, z-score = -1.34, R) >dp4.chrXR_group8 7794086 105 + 9212921 CCGACAGCAAGAGUAGUAGAUUAAAGGGCCAACGCGUCAUGGCCCCUUAGGCCGGGGCCGUUCAGCGCGACGCCUAUGGGUCCCAACAGAUUCUCACACUGCAAC---------- ............(((((........((((...(((((.((((((((.......))))))))...)))))..))))..(((((......)))))....)))))...---------- ( -36.70, z-score = -1.45, R) >droPer1.super_48 574931 105 + 581423 CCGACAGCAAGAGUAGUAGAUUAAAGGGCCAACGCGUCAUGGCCCCUUAGGCCGGGGCCGUUCAGCGCGACGCCUAUGGGUCCCAACAGAUUCUCACACUGCAAC---------- ............(((((........((((...(((((.((((((((.......))))))))...)))))..))))..(((((......)))))....)))))...---------- ( -36.70, z-score = -1.45, R) >droWil1.scaffold_180949 4861653 115 + 6375548 CCGAUAGCAAUAGUAAUAAAUUAAAGGGCCAACGCGUCAUGGCCCCUUAGGCCGGGGCCGUUCAGCGCGACGCCUAUGGUUCCAAACAGAUUUACAAGCUGCAAAAUGGAAAAAA (((...(((.(.((((.........((((((..(((((..((((((.......))))))((.....)))))))...)))))).........)))).)..)))....)))...... ( -33.47, z-score = -0.76, R) >droMoj3.scaffold_6680 12266275 109 + 24764193 CCGAGAGCAAUAGUAGUAAAUUGAAGGGCCAACGCGUCAUGGCCCCUUAGGCCGGGGCCGUUCAGCGCGACGCCUAUGGGUGCUAACAGAUUUACACGCUGCAAAUAUA------ ......(((...((.(((((((....(((...(((((.((((((((.......))))))))...)))))..(((....))))))....)))))))))..))).......------ ( -39.50, z-score = -1.58, R) >droVir3.scaffold_13049 6267633 109 - 25233164 CCGAGAGCAAUAGUAGUAAAUUGAAGGGCCAACGCGUCAUGGCCCCUUAGGCCGGGGCCGUUCAGCGCGACGCCUAUGGGUGCUAACAGAUUUACACGCUGCAAGGAUA------ ((....(((...((.(((((((....(((...(((((.((((((((.......))))))))...)))))..(((....))))))....)))))))))..)))..))...------ ( -40.10, z-score = -1.45, R) >droGri2.scaffold_15110 7902726 114 - 24565398 CCGAGAGCAAUAGUAGUAAAUUGAAGGGCCAACGCGUCAUGGCCCCUUAGGCCGGGGCCGUUCAGCGCGACGCCUAUGGGUGCUAACAGAUUUACAAGCUGCAAGAAGAACACA- ......(((......(((((((....(((...(((((.((((((((.......))))))))...)))))..(((....))))))....)))))))....)))............- ( -36.70, z-score = -0.77, R) >anoGam1.chr3L 3742159 101 - 41284009 CCGAGAGCAGUGCAAGCAGUAGGACGGGCCAACGCGUCAUGGCCCCCUCGAAUGGGGCCGAUCAGCGCGACUCCAACCGAUUUCGA---ACACCGGCUGUGGAA----------- ..(.(((..((((.........((((.(....).)))).(((((((.......)))))))....))))..))))..(((...(((.---....)))...)))..----------- ( -34.20, z-score = 0.37, R) >consensus CCGACAGCAAGAGUAGUAGAUUAAAGGGCCAACGCGUCAUGGCCCCUUAGGCCGGGGCCGUUCAGCGCGACGCCUAUGGGUCCCAACAGAUUCUCACACUGCAAUGA________ ......(((................((((...(((((.((((((((.......))))))))...)))))..)))).(((...)))..............)))............. (-29.39 = -29.50 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:22 2011