
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,719,436 – 19,719,556 |
| Length | 120 |
| Max. P | 0.781048 |

| Location | 19,719,436 – 19,719,556 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.18 |
| Shannon entropy | 0.44109 |
| G+C content | 0.55056 |
| Mean single sequence MFE | -46.60 |
| Consensus MFE | -18.61 |
| Energy contribution | -17.58 |
| Covariance contribution | -1.03 |
| Combinations/Pair | 1.77 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
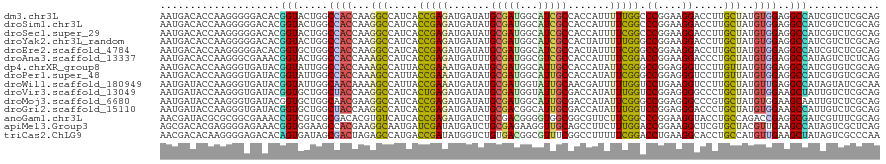
>dm3.chr3L 19719436 120 - 24543557 AAUGACACCAAGGGGGACACGGUACUGGCCACCAAGGCCAUCACCGAGAUGAUAUGCGAUGGCAUCGCCACCAUUUUUGGCCCGGAAGGACCUUGCUAUGUGGAGGCCAUCGUCUCGCAG ...........(.(((((.((((..(((((.....)))))..))))...........((((((....((((.......((.((....)).)).......))))..))))))))))).).. ( -48.94, z-score = -2.73, R) >droSim1.chr3L 19045643 120 - 22553184 AAUGACACCAAGGGGGACACGGUACUGGCCACCAAGGCCAUCACCGAGAUGAUAUGCGAUGGCAUCGCCACCAUUUUCGGCCCGGAAGGACCUUGCUAUGUGGAGGCCAUCGUCUCGCAG ...........(.(((((.((((..(((((.....)))))..))))...........((((((....((((.......((.((....)).)).......))))..))))))))))).).. ( -48.24, z-score = -2.47, R) >droSec1.super_29 417107 120 - 700605 AAUGACACCAAGGGGGACACGGUACUGGCCACCAAGGCCAUCACCGAGAUGAUAUGCGAUGGCAUCGCCACCAUUUUCGGCCCGGAAGGACCUUGCUAUGUGGAGGCCAUCGUCUCGCAG ...........(.(((((.((((..(((((.....)))))..))))...........((((((....((((.......((.((....)).)).......))))..))))))))))).).. ( -48.24, z-score = -2.47, R) >droYak2.chr3L_random 3348184 120 - 4797643 AAUGACACCAAGGGGGACACGGUACUGGCCACCAAGGCCAUCACCGAGAUGAUAUGCGAUGGCAUCGCCACUAUUUUUGGGCCGGAAGGACCUUGCUAUGUGGAGGCCAUCGUCUCGCAG ...........(.(((((.((((..(((((.....)))))..))))...........((((((....((((.......(((((....)).)))......))))..))))))))))).).. ( -50.62, z-score = -3.37, R) >droEre2.scaffold_4784 19511940 120 - 25762168 AAUGACACCAAGGGGGACACGGUGCUGGCCACCAAGGCCAUCACCGAGAUGAUAUGCGAUGGCAUCGCCACUAUUUUCGGGCCGGAAGGACCUUGCUAUGUGGAGGCCAUCGUCUCGCAG ...........(.(((((.(((((.(((((.....))))).)))))...........((((((....((((.......(((((....)).)))......))))..))))))))))).).. ( -53.42, z-score = -3.53, R) >droAna3.scaffold_13337 6116054 120 + 23293914 AAUGACACCAAGGGCGAAACGGUACUGGCCACCAAAGCCAUCACCGAGAUGAUUUGCGAUGGCGUCGCCACCAUUUUCGGACCGGAAGGACCCUGCUAUGUGGAGGCCAUAGUCUCUCAG ..((((((..(((((((((.(((...(((.((....((((((.(.((.....)).).)))))))).))))))..)))))..((....)).)))).....)))(((((....)))))))). ( -37.50, z-score = -0.51, R) >dp4.chrXR_group8 7793711 120 - 9212921 AAUGACACCAAGGGUGAUACGGUAUUGGCCACCAAAGCCAUUACCGAAAUGAUAUGCGAUGGCAUUGCCACCAUAUUCGGGCCGGAGGGUCCUUGUUAUGUGGAGGCCAUCGUGUCGCAG .....((((...))))...(((((.((((.......)))).)))))...((..((((((((((....((((...((..(((((....)))))..))...))))..))))))))))..)). ( -48.20, z-score = -2.73, R) >droPer1.super_48 574556 120 - 581423 AAUGACACCAAGGGUGAUACGGUAUUGGCCACCAAAGCCAUUACCGAAAUGAUAUGCGAUGGCAUUGCCACCAUAUUCGGGCCGGAGGGUCCUUGUUAUGUGGAGGCCAUCGUGUCGCAG .....((((...))))...(((((.((((.......)))).)))))...((..((((((((((....((((...((..(((((....)))))..))...))))..))))))))))..)). ( -48.20, z-score = -2.73, R) >droWil1.scaffold_180949 4861288 120 - 6375548 AAUGAUACCAAGGGUGAUACGGUAUUGGCAACAAAAGCCAUUACCGAAAUGAUAUGCGAUGGUAUUGCAACGAUUUUUGGUCCUGAAGGUCCUUGCUAUGUUGAGGCCAUAGUAUCGCAA .....((((...))))...(((((.((((.......)))).)))))........(((((((.(((.((..((((....((.((....)).)).......))))..)).))).))))))). ( -37.60, z-score = -2.37, R) >droVir3.scaffold_13049 6266562 120 + 25233164 AAUGAUACCAAGGGUGAUACGGUGCUGGCUACCAAGGCCAUCACUGAGAUGAUAUGCGAUGGUAUUGCGACCAUAUUUGGUCCGGAGGGGCCCUGCUAUGUGGAAGCUAUUGUCUCGCAG .....((((...))))...(((((.(((((.....))))).)))))........(((((.(..(..((..((((((..(((((....))))).....))))))..))...)..)))))). ( -40.70, z-score = -0.93, R) >droMoj3.scaffold_6680 12265557 120 - 24764193 AAUGAUACCAAGGGUGAUACGGUGCUGGCAACGAAGGCCAUCACCGAGAUGAUAUGCGAUGGCAUUGCGACCAUAUUCGGGCCGGAGGGCCCGUGCUAUGUGGAAGCAAUUGUCUCGCAG .....((((...))))...(((((.((((.......)))).)))))........(((((.((((((((..((((((.((((((....))))))....))))))..)))).))))))))). ( -53.40, z-score = -4.01, R) >droGri2.scaffold_15110 7901839 120 + 24565398 AAUGAUACCAAGGGUGAUACGGUGCUGGCUACCAAGGCCAUCACCGAGAUGAUAUGCGACGGCAUUGCGACCAUAUUUGGUCCGGAGGGACCCUGCUAUGUGGAAGCCAUUGUCUCGCAG .....((((...))))...(((((.(((((.....))))).)))))........(((((.((((..((..((((((..(((((....))))).....))))))..))...))))))))). ( -46.70, z-score = -2.55, R) >anoGam1.chr3L 3741557 120 + 41284009 AACGAUACGCGCGGCGAAACCGUCGUCGCGACACGUGUCAUCACCGAGAUGAUCUGCGACGGGGUGGCGGCGUUCUUCGGCCCGGAAGGUACCUGCCAGACCGAGGCGAUCGUUUCGCAG ...((((((((((((((.....))))))))...)))))).....((((((((((.((..(((..((((((.(((((((......))))).))))))))..)))..))))))))))))... ( -57.80, z-score = -2.52, R) >apiMel3.Group3 4730684 120 + 12341916 AGCGACACGAGGGGAGAAACGGUGGAAGCCACGAAGGCAAUGAUCGAUAUGAUCUGCGAGAAGGUUGCAGCCUUCUUUGGACCGGAAGGCUCGUGCUACGUUGAAGCCAUAGUCGCUCAG (((((((((((..........((((...))))((((((...((((.....))))(((((.....)))))))))))......((....)))))))(((.......)))....))))))... ( -40.50, z-score = -1.01, R) >triCas2.ChLG9 489639 120 + 15222296 AACGACACAAGGGGAGACACAGUGAUAGCGACUAGAGCCAUGACCGAUAUGGUCUGUGACGGCGUUUCGGCCUUUUUCGGACCUGAAGGCACCUGCCAUGUUGAAGCUAUAGUCGCCCAA .....(((....(....)...)))...(((((((.(((......((((((((((((.((.(((......))).))..))))))....(((....)))))))))..))).))))))).... ( -39.00, z-score = -1.33, R) >consensus AAUGACACCAAGGGUGACACGGUACUGGCCACCAAGGCCAUCACCGAGAUGAUAUGCGAUGGCAUUGCCACCAUUUUCGGGCCGGAAGGACCUUGCUAUGUGGAGGCCAUCGUCUCGCAG ....................(((.....((((...(((.....(((((.......(((((...))))).......))))).((....)).....)))..))))..)))............ (-18.61 = -17.58 + -1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:20 2011