| Sequence ID | dm3.chr3L |
|---|---|
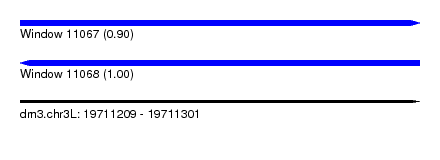
| Location | 19,711,209 – 19,711,301 |
| Length | 92 |
| Max. P | 0.999272 |

| Location | 19,711,209 – 19,711,301 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 67.55 |
| Shannon entropy | 0.65214 |
| G+C content | 0.48382 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -14.87 |
| Energy contribution | -14.92 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
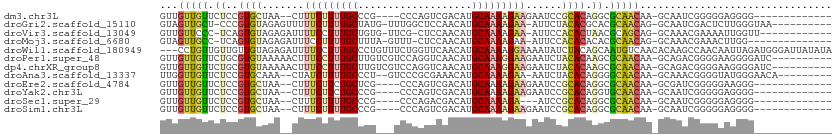
>dm3.chr3L 19711209 92 + 24543557 GUUGUUGUUCUCCGUGCUAA--CUUUCUUUUGCCCG----CCCAGUCGACAUGCAAAAGAAGAAUCCGCACAGGCGCAACAA-GCAAUCGGGGGAGGGG------------- ....((.(((((((((((..--.((((((((((.((----......))....))))))))))....(((....))).....)-)))..))))))).)).------------- ( -24.80, z-score = 0.59, R) >droGri2.scaffold_15110 15532711 98 + 24565398 GUAGUUGCU-CCCGUGUAGAGUUUUUCUUUUGCUAUG-UUUGGCUCCAACAUGCAAAAGAA-AUUCUACACGCACGCAACAG-GCAAUCGACUCUUGGGUAA---------- ...(((((.-..(((((((((((..((((((((.(((-((.(....)))))))))))))))-))))))))))...)))))..-.......(((....)))..---------- ( -36.20, z-score = -4.72, R) >droVir3.scaffold_13049 11529057 95 - 25233164 GUUGUUCCC-UCAGUGUAGAGAUUUUCCUUUGCUGUG-UUCG-CUCCAACAUGCAAAAGAA-AUUCCACACUAACGCAGCAG-GCAAACGAAAAUUGGUU------------ (((((....-..(((((.(.((.((((.(((((.(((-((..-....)))))))))).)))-).))))))))...)))))..-.................------------ ( -21.00, z-score = -0.64, R) >droMoj3.scaffold_6680 13642634 93 + 24764193 GUAGUUGCC-UCAGUGUAGAGAUUUCCUUUUGCUUUA-GUUU-CUCCAACAUGCAAAAGAA-AUUCCACACACACGCAACAG-GCAAACGAAACUUGG-------------- ...(((((.-...((((.(..(((((.((((((....-(((.-....)))..)))))))))-))..)))))....)))))..-...............-------------- ( -21.10, z-score = -1.65, R) >droWil1.scaffold_180949 4853084 109 + 6375548 ---CCUGUUGUUGUUGUAGAGAUUUUCCUUUGCCCUGUUUCUGGUUCAACAUGCAAAAGGAAAAUAUCUACAGCAAUGUCAACACAAGCCAACAAUUAGAUGGGAUUAUAUA ---..(((((((((((((((.((((((((((((..((((........)))).)).)))))))))).))))))))))...)))))....(((.(.....).)))......... ( -31.70, z-score = -3.13, R) >droPer1.super_48 566602 101 + 581423 GUUGUUGUUCUGCGUGUAAAAACUUUCCUUUGCUUGUCGUCCAGGUCAACAUGCAAAGGAAGAAUCUACACAAGCGCAACAA-GCAGACGGGGAAGGGGAUC---------- .(((((((.((..(((((.....((((((((((.(((.(.......).))).))))))))))....))))).)).)))))))-...................---------- ( -28.60, z-score = -1.13, R) >dp4.chrXR_group8 7785194 101 + 9212921 GUUGUUGUUCUGCGUGUAAAAACUUUCCUUUGCUUGUCGUCCAGGUCAACAUGCAAAGGAAGAAUCUACACAAGCGCAACAA-GCAGACGGGGAAGGGGAUC---------- .(((((((.((..(((((.....((((((((((.(((.(.......).))).))))))))))....))))).)).)))))))-...................---------- ( -28.60, z-score = -1.13, R) >droAna3.scaffold_13337 6109105 97 - 23293914 UUGGUUGUUCUCCGUGCAAA--CUAUCUUUUGCCCU--GUCCCGCGAAACAUGCAAAAGAA-AAUCUACACAGGGGCAACAA-GCAAACGGGGUAUGGGAACA--------- .....(((((.((((((...--...((.(((((..(--((((((((.....)))...((..-...)).....))))))....-))))).)).)))))))))))--------- ( -27.30, z-score = -0.97, R) >droEre2.scaffold_4784 19504142 92 + 25762168 GUUGUUGUUCUCCGUGCUAA--CUUUCUUCUGCUCG----CCCAGUCGACAUGCAAAAGAAGAAUCCGCACAGGCGCAACAA-GCGAUCGGGGGAAGGG------------- .....(.((((((((((...--..((((((((((((----......)))...))...)))))))...)))).(((((.....-))).)).)))))).).------------- ( -25.10, z-score = 0.39, R) >droYak2.chr3L 3381832 92 - 24197627 GUUGUUGUUCUCCGUGCUAA--CUUUCUUCUGCCCG----CCCAGUCGACAUGCAAAAGAAGAAUCCGCACAGGUGCAACAA-GCAAUCGGGGGAGGGG------------- ....((.(((((((((((..--..(((((((....(----(...........))...)))))))...(((....)))....)-)))..))))))).)).------------- ( -23.50, z-score = 0.95, R) >droSec1.super_29 409241 89 + 700605 GUUGUUGUUCUCCGUGCUAA--CUUUCUUUUGCCCG----CCCAGACGACAUGCAAAAGA---AUCCGCACAGGCGCAACAA-GCAAUCGGGGGAGGGG------------- ....((.(((((((((((..--..(((((((((.((----......))....))))))))---)..(((....))).....)-)))..))))))).)).------------- ( -24.70, z-score = 0.36, R) >droSim1.chr3L 19037288 92 + 22553184 GUUGUUGUUCUCCGUGCUAA--CUUUCUUUUGCCCG----CCCAGUCGACAUGCAAAAGAAGAAUCCGCACAGGCGCAACAA-GCAAUCGGGGGAGGGG------------- ....((.(((((((((((..--.((((((((((.((----......))....))))))))))....(((....))).....)-)))..))))))).)).------------- ( -24.80, z-score = 0.59, R) >consensus GUUGUUGUUCUCCGUGCAAA__CUUUCUUUUGCCCG___UCCCAGUCAACAUGCAAAAGAAGAAUCCACACAGGCGCAACAA_GCAAUCGGGGGAGGGG_____________ ...(((((.((..((((.......(((((((((...................)))))))))......)))).)).)))))................................ (-14.87 = -14.92 + 0.05)

| Location | 19,711,209 – 19,711,301 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 67.55 |
| Shannon entropy | 0.65214 |
| G+C content | 0.48382 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -14.27 |
| Energy contribution | -14.19 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
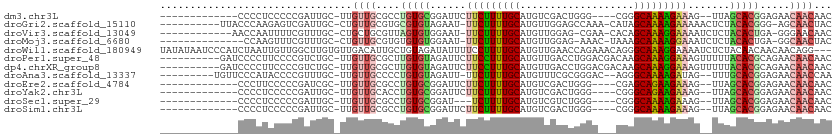
>dm3.chr3L 19711209 92 - 24543557 -------------CCCCUCCCCCGAUUGC-UUGUUGCGCCUGUGCGGAUUCUUCUUUUGCAUGUCGACUGGG----CGGGCAAAAGAAAG--UUAGCACGGAGAACAACAAC -------------................-((((((..(((((((.((((.(((((((((.((((.....))----)).)))))))))))--)).)))))).)..)))))). ( -32.30, z-score = -2.04, R) >droGri2.scaffold_15110 15532711 98 - 24565398 ----------UUACCCAAGAGUCGAUUGC-CUGUUGCGUGCGUGUAGAAU-UUCUUUUGCAUGUUGGAGCCAAA-CAUAGCAAAAGAAAAACUCUACACGGG-AGCAACUAC ----------...................-..(((((.(.((((((((.(-(((((((((((((((....).))-))).))))))))))...)))))))).)-.)))))... ( -35.00, z-score = -4.41, R) >droVir3.scaffold_13049 11529057 95 + 25233164 ------------AACCAAUUUUCGUUUGC-CUGCUGCGUUAGUGUGGAAU-UUCUUUUGCAUGUUGGAG-CGAA-CACAGCAAAGGAAAAUCUCUACACUGA-GGGAACAAC ------------.................-.((.(.(.((((((((((.(-(((((((((.((((....-..))-))..))))))))))...))))))))))-.).).)).. ( -27.50, z-score = -1.86, R) >droMoj3.scaffold_6680 13642634 93 - 24764193 --------------CCAAGUUUCGUUUGC-CUGUUGCGUGUGUGUGGAAU-UUCUUUUGCAUGUUGGAG-AAAC-UAAAGCAAAAGGAAAUCUCUACACUGA-GGCAACUAC --------------...............-..(((((.(..(((((((.(-(((((((((...((((..-...)-))).))))))))))...)))))))..)-.)))))... ( -28.40, z-score = -2.63, R) >droWil1.scaffold_180949 4853084 109 - 6375548 UAUAUAAUCCCAUCUAAUUGUUGGCUUGUGUUGACAUUGCUGUAGAUAUUUUCCUUUUGCAUGUUGAACCAGAAACAGGGCAAAGGAAAAUCUCUACAACAACAACAGG--- .........((.......(((..((....))..)))(((.((((((.((((((((((.((.((((........))))..)))))))))))).)))))).))).....))--- ( -32.80, z-score = -3.82, R) >droPer1.super_48 566602 101 - 581423 ----------GAUCCCCUUCCCCGUCUGC-UUGUUGCGCUUGUGUAGAUUCUUCCUUUGCAUGUUGACCUGGACGACAAGCAAAGGAAAGUUUUUACACGCAGAACAACAAC ----------...................-((((((..((((((((((..((((((((((.(((((.......))))).))))))).)))..)))))))).))..)))))). ( -34.50, z-score = -4.53, R) >dp4.chrXR_group8 7785194 101 - 9212921 ----------GAUCCCCUUCCCCGUCUGC-UUGUUGCGCUUGUGUAGAUUCUUCCUUUGCAUGUUGACCUGGACGACAAGCAAAGGAAAGUUUUUACACGCAGAACAACAAC ----------...................-((((((..((((((((((..((((((((((.(((((.......))))).))))))).)))..)))))))).))..)))))). ( -34.50, z-score = -4.53, R) >droAna3.scaffold_13337 6109105 97 + 23293914 ---------UGUUCCCAUACCCCGUUUGC-UUGUUGCCCCUGUGUAGAUU-UUCUUUUGCAUGUUUCGCGGGAC--AGGGCAAAAGAUAG--UUUGCACGGAGAACAACCAA ---------(((((((...........((-.....))....(((((((((-.((((((((.(((((....))))--)..)))))))).))--))))))))).)))))..... ( -34.30, z-score = -3.42, R) >droEre2.scaffold_4784 19504142 92 - 25762168 -------------CCCUUCCCCCGAUCGC-UUGUUGCGCCUGUGCGGAUUCUUCUUUUGCAUGUCGACUGGG----CGAGCAGAAGAAAG--UUAGCACGGAGAACAACAAC -------------................-((((((..(((((((.((((.(((((((((.((((.....))----)).)))))))))))--)).)))))).)..)))))). ( -32.40, z-score = -2.30, R) >droYak2.chr3L 3381832 92 + 24197627 -------------CCCCUCCCCCGAUUGC-UUGUUGCACCUGUGCGGAUUCUUCUUUUGCAUGUCGACUGGG----CGGGCAGAAGAAAG--UUAGCACGGAGAACAACAAC -------------................-((((((..(((((((.((((.(((((((((.((((.....))----)).)))))))))))--)).)))))).)..)))))). ( -31.10, z-score = -1.75, R) >droSec1.super_29 409241 89 - 700605 -------------CCCCUCCCCCGAUUGC-UUGUUGCGCCUGUGCGGAU---UCUUUUGCAUGUCGUCUGGG----CGGGCAAAAGAAAG--UUAGCACGGAGAACAACAAC -------------................-((((((..(((((((...(---((((((((.((((.....))----)).)))))))))..--...)))))).)..)))))). ( -30.10, z-score = -1.36, R) >droSim1.chr3L 19037288 92 - 22553184 -------------CCCCUCCCCCGAUUGC-UUGUUGCGCCUGUGCGGAUUCUUCUUUUGCAUGUCGACUGGG----CGGGCAAAAGAAAG--UUAGCACGGAGAACAACAAC -------------................-((((((..(((((((.((((.(((((((((.((((.....))----)).)))))))))))--)).)))))).)..)))))). ( -32.30, z-score = -2.04, R) >consensus _____________CCCCUCCCCCGAUUGC_UUGUUGCGCCUGUGUGGAUUCUUCUUUUGCAUGUUGACUGGGA___CGAGCAAAAGAAAG__UUUACACGGAGAACAACAAC ................................((((....(((((.((...(((((((((...................)))))))))....)).))))).....))))... (-14.27 = -14.19 + -0.08)
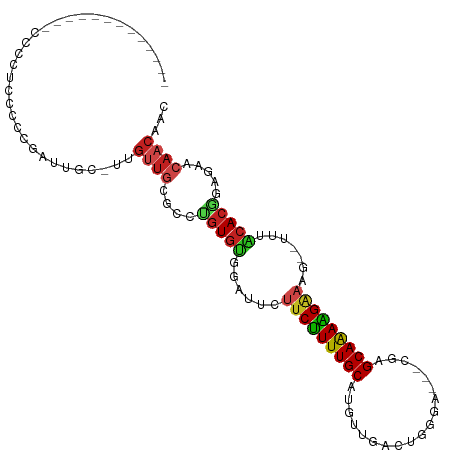
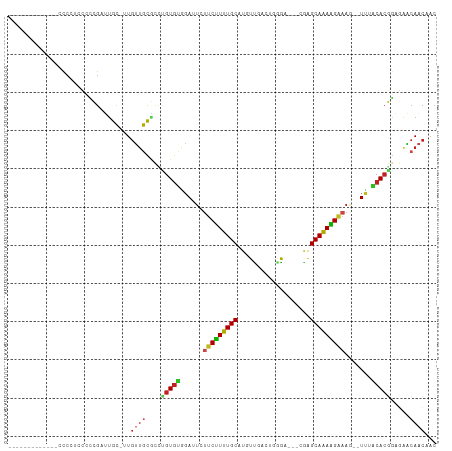
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:19 2011