
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,682,520 – 19,682,604 |
| Length | 84 |
| Max. P | 0.637555 |

| Location | 19,682,520 – 19,682,604 |
|---|---|
| Length | 84 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 69.60 |
| Shannon entropy | 0.54593 |
| G+C content | 0.49927 |
| Mean single sequence MFE | -18.18 |
| Consensus MFE | -8.75 |
| Energy contribution | -10.17 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.637555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
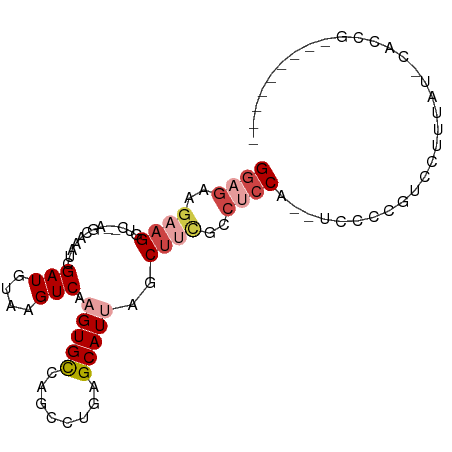
>dm3.chr3L 19682520 84 - 24543557 GGAGAAGAAGGUC--AGCAAAUCGAUGUAAGUCAAGUGCCAGCCUGAGCAUUAGCUUCGCCUCCAUUUCCCCGUUCUUUAU-CACCG--------- (((((.(.((((.--(((.....(((....))).((((((.....).))))).)))..)))).).)))))...........-.....--------- ( -18.20, z-score = -0.92, R) >droSim1.chr3L 19010923 82 - 22553184 GGAGAAGAAGGUC--AGCAAAUCGAUGUAAGUCAAGUGCCAGCCUGAGCAUUAGCUUUGCCUCCA--UCCCCGUUCUUUAU-CACCG--------- ((((....((((.--.(((....(((....)))...)))..))))((((....))))...)))).--..............-.....--------- ( -17.90, z-score = -0.79, R) >droSec1.super_29 385050 82 - 700605 GGAGAAGAAGGUC--AGCAAAUCGAUGUUAGUCAAGUGCCAGCCUGAGCAUAAGCUUCGCCUCCA--UCCCCGUUCUUUAU-CAUCG--------- ((((....((((.--.(((....(((....)))...)))..))))((((....))))...)))).--..............-.....--------- ( -17.90, z-score = -0.81, R) >droYak2.chr3L 3350599 94 + 24197627 GGAGAAGAAGGUC--AACAAAUCGAUGUAAGUCAAGUGCCAGCCUGAGCAUUAGCUUCUCCUCCAUCCCCCUCCCGCAUACGUAUUCGUAUCAACG ((((.(((((...--........(((....))).((((((.....).)))))..))))).))))...........(.(((((....)))))).... ( -21.00, z-score = -2.08, R) >droEre2.scaffold_4784 19480131 92 - 25762168 GGAGAAGAAGGUC--AGCAAAUCGAUGUAAGUCAAGUGCCAGCCUGAGCAUUAGCUGCUCCCCCA-GCCCCUCCCGUUCAC-CACCCGUAUCAACG ((((....((((.--.(((....(((....)))...)))..))))(((((.....))))).....-....)))).......-.............. ( -20.00, z-score = -1.29, R) >droAna3.scaffold_13337 6082487 79 + 23293914 GAAGAGACAGACCGAAAAAAAUCGAUAUAAGUCAAGUGUCAAGUGUGGCAUGAACAAAGUGCCCCAAUUCCCAUCUCCU----------------- ...((((..((((((......))(((....)))..).)))...((.(((((.......))))).)).......))))..----------------- ( -14.10, z-score = -1.74, R) >consensus GGAGAAGAAGGUC__AGCAAAUCGAUGUAAGUCAAGUGCCAGCCUGAGCAUUAGCUUCGCCUCCA__UCCCCGUCCUUUAU_CACCG_________ ((((..((((.............(((....))).(((((........)))))..))))..))))................................ ( -8.75 = -10.17 + 1.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:17 2011