
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,667,849 – 19,668,043 |
| Length | 194 |
| Max. P | 0.993383 |

| Location | 19,667,849 – 19,667,914 |
|---|---|
| Length | 65 |
| Sequences | 6 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 66.52 |
| Shannon entropy | 0.61102 |
| G+C content | 0.59336 |
| Mean single sequence MFE | -19.53 |
| Consensus MFE | -16.12 |
| Energy contribution | -16.28 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
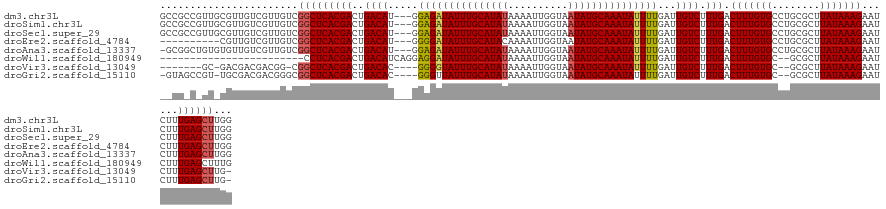
>dm3.chr3L 19667849 65 - 24543557 -------------AAAGGCAGCGACGUCGCUUUGGCUAGGACUGUCUUGGCCAACUACCCCCCCCCCCUCCUUCUCAC---- -------------.((((.((((....))))(((((((((.....)))))))))...............)))).....---- ( -18.60, z-score = -1.64, R) >droSim1.chr3L 18995604 66 - 22553184 -------------AAAGGCAGCGACGUCGCUUUGGCUAGGACUGUCUUGGCCAACUACCCCGCAUUCCCCCCCUCUUGC--- -------------....((((((....))))(((((((((.....))))))))).......))................--- ( -19.00, z-score = -0.91, R) >droSec1.super_29 370458 65 - 700605 -------------AAAGGCAGCGACGUCGCUUUGGCUAGGACUGUCUUGGCCAACUACCCCGCACU-CCCCCCUCUUAC--- -------------....((((((....))))(((((((((.....))))))))).......))...-............--- ( -19.00, z-score = -1.40, R) >droYak2.chr3L 3321499 78 + 24197627 -AAAGGCAGCAACAGCGACAGCGACGUCGCUUUGGCUAGGACUGUCUUGGCCAACUUCCCCCCCCCCCCCUUUUCCCCU--- -(((((.......((((((......))))))(((((((((.....)))))))))..............)))))......--- ( -23.60, z-score = -1.97, R) >droEre2.scaffold_4784 19466220 54 - 25762168 ------------------CAGCGACGUCGCUUUGGCUAGGACUGUCUUGGCCAACUUCGCCCCCCCUUCUUC---------- ------------------.((((....))))(((((((((.....)))))))))..................---------- ( -16.50, z-score = -1.86, R) >droAna3.scaffold_13337 6068938 82 + 23293914 AAUGGCAAAAGGCGACUUCAGCGACGUCGCUUUGGCUAGGACUGUCUCGGCCAAGUUCCCCUUUAUACUACCAACUUCUACC ..(((...((((((((.((...)).))))))))(((..((.....))..)))..................)))......... ( -20.50, z-score = -0.84, R) >consensus _____________AAAGGCAGCGACGUCGCUUUGGCUAGGACUGUCUUGGCCAACUACCCCCCCCCCCCCCCCUCUUC____ ...................((((....))))(((((((((.....)))))))))............................ (-16.12 = -16.28 + 0.17)

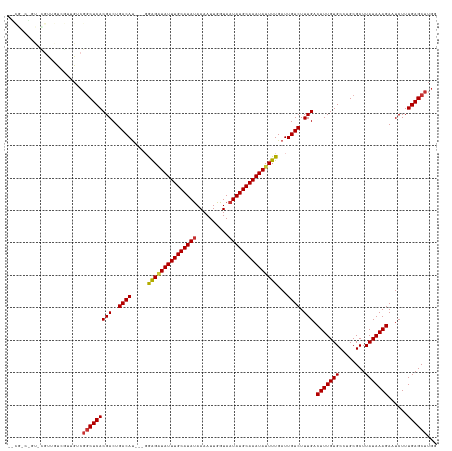
| Location | 19,667,914 – 19,668,043 |
|---|---|
| Length | 129 |
| Sequences | 8 |
| Columns | 132 |
| Reading direction | forward |
| Mean pairwise identity | 85.01 |
| Shannon entropy | 0.30193 |
| G+C content | 0.40252 |
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -27.82 |
| Energy contribution | -27.86 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.981817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19667914 129 + 24543557 GCCGCCGUUGCGUUGUCGUUGUCGGCUCACGACUGACAU---GGAGAUAUUUGCAUAUAAAAUUGGUAAUAUGCAAAUAUUUUGAUUGUCUUUGACUUUGUGCCUGCGCUUAUAAAGAAUCUUUGAGCUUGG .((((.(..(((..((((..((((.....)))).(((((---.(((((((((((((((..........))))))))))))))).).))))..))))..)))..).))(((((...........)))))..)) ( -38.70, z-score = -2.82, R) >droSim1.chr3L 18995670 129 + 22553184 GCCGCCGUUGCGUUGUCGUUGUCGGCUCACGACUGACAU---GGAGAUAUUUGCAUAUAAAAUUGGUAAUAUGCAAAUAUUUUGAUUGUCUUUGACUUUGUGCCUGCGCUUAUAAAGAAUCUUUGAGCUUGG .((((.(..(((..((((..((((.....)))).(((((---.(((((((((((((((..........))))))))))))))).).))))..))))..)))..).))(((((...........)))))..)) ( -38.70, z-score = -2.82, R) >droSec1.super_29 370523 129 + 700605 GCCGCCGUUGCGUUGUCGUUGUCGGCUCACGACUGACAU---GGAGAUAUUUGCAUAUAAAAUUGGUAAUAUGCAAAUAUUUUGAUUGUCUUUGACUUUGUGCCUGCGCUUAUAAAGAAUCUUUGAGCUUGG .((((.(..(((..((((..((((.....)))).(((((---.(((((((((((((((..........))))))))))))))).).))))..))))..)))..).))(((((...........)))))..)) ( -38.70, z-score = -2.82, R) >droEre2.scaffold_4784 19466274 119 + 25762168 ----------CGUUGUCGUUGUCGGCUCACGACUGACAU---GGGGAUAUUUGCAUACAAAAUUGGUAAUAUGCAAAUAUUUUGAUUGUCUUUGACUUUGUGCCUGCGCUUAUAAAGAAUCUUUGAGCUUGG ----------((..((((..((((.....)))).(((((---.((((((((((((((............)))))))))))))).).))))..))))..))..((...(((((...........)))))..)) ( -29.40, z-score = -1.32, R) >droAna3.scaffold_13337 6069020 128 - 23293914 -GCGGCUGUGUGUUGUCGUUGUCGGCUCACGACUGACAU---GGAGAUAUUUGCAUAUAAAAUUGGUAAUAUGCAAAUAUUUUGAUUGUCUUUGACUUUGUGCCUGCGCUUAUAAAGAAUCUUUGAGCUUGG -((((..(..((..((((..((((.....)))).(((((---.(((((((((((((((..........))))))))))))))).).))))..))))..))..)))))(((((...........))))).... ( -38.60, z-score = -3.08, R) >droWil1.scaffold_180949 4801766 106 + 6375548 ------------------------CCUCACGACUGACAUCAGGAGGAUAUUUGCAUAUAAAAUUGGUAAUAUGCAAAUAUUUUGAUUGUCUUUGACUUUGUGC--GCGCUUAUAAAGAAUCUUUGAGCUUUG ------------------------.(((((((..((((((((...(((((((((((((..........))))))))))))))))).)))).))).(((((((.--.....)))))))......))))..... ( -25.40, z-score = -2.14, R) >droVir3.scaffold_13049 13253183 116 + 25233164 -------GC-GACGACGACGG-CGGCUCACGACUGACAC----GGGGUAUUUGCAUAUAAAAUUGGUAAUAUGCAAAUAUUUUGAUUGUCUUUGACUUUGUGC--GCGCUUAUAAAGAAUCUUUGAGCUUG- -------.(-(.((....)).-))((((((((..(((((----(((((((((((((((..........))))))))))))))))..)))).))).(((((((.--.....)))))))......)))))...- ( -31.10, z-score = -2.07, R) >droGri2.scaffold_15110 18693088 123 + 24565398 -GUAGCCGU-UGCGACGACGGGCGGCUCACGACUGACAC----GGGUUAUUUGCAUAUAAAAUUGGUAAUAUGCAAAUAUUUUGAUUGUCUUUGACUUUGUGC--GCGCUUAUAAAGAAUCUUUGAGCUUG- -....((((-((...))))))..(((((((((..(((((----(((.(((((((((((..........))))))))))).))))..)))).))).(((((((.--.....)))))))......))))))..- ( -33.30, z-score = -1.37, R) >consensus __CG_C_GU_CGUUGUCGUUGUCGGCUCACGACUGACAU___GGAGAUAUUUGCAUAUAAAAUUGGUAAUAUGCAAAUAUUUUGAUUGUCUUUGACUUUGUGCCUGCGCUUAUAAAGAAUCUUUGAGCUUGG .......................(((((((((..((((.....(((((((((((((((..........)))))))))))))))...)))).))).(((((((........)))))))......))))))... (-27.82 = -27.86 + 0.05)
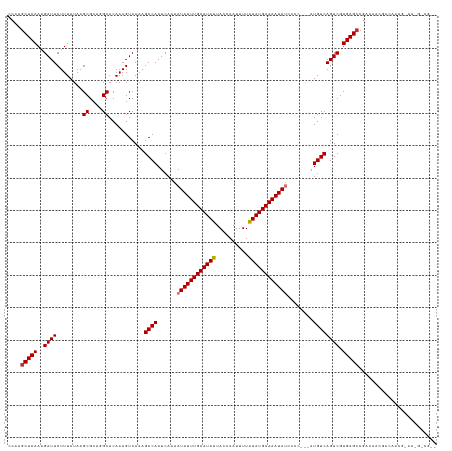
| Location | 19,667,914 – 19,668,043 |
|---|---|
| Length | 129 |
| Sequences | 8 |
| Columns | 132 |
| Reading direction | reverse |
| Mean pairwise identity | 85.01 |
| Shannon entropy | 0.30193 |
| G+C content | 0.40252 |
| Mean single sequence MFE | -26.29 |
| Consensus MFE | -22.39 |
| Energy contribution | -22.65 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.991388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
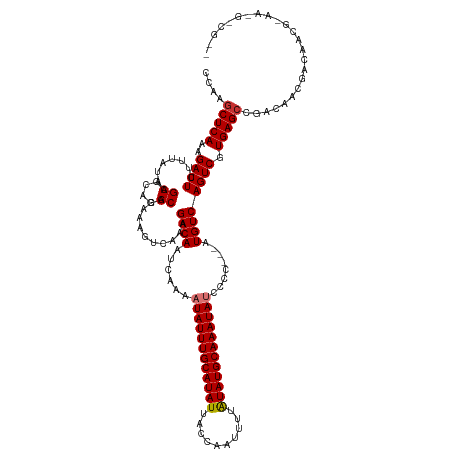
>dm3.chr3L 19667914 129 - 24543557 CCAAGCUCAAAGAUUCUUUAUAAGCGCAGGCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUAUCUCC---AUGUCAGUCGUGAGCCGACAACGACAACGCAACGGCGGC ((..(((.((((...))))...)))((..((...........((((......((((((((((((..........))))))))))))....---.)))).(((((.........)))))...))....)))). ( -28.90, z-score = -2.37, R) >droSim1.chr3L 18995670 129 - 22553184 CCAAGCUCAAAGAUUCUUUAUAAGCGCAGGCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUAUCUCC---AUGUCAGUCGUGAGCCGACAACGACAACGCAACGGCGGC ((..(((.((((...))))...)))((..((...........((((......((((((((((((..........))))))))))))....---.)))).(((((.........)))))...))....)))). ( -28.90, z-score = -2.37, R) >droSec1.super_29 370523 129 - 700605 CCAAGCUCAAAGAUUCUUUAUAAGCGCAGGCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUAUCUCC---AUGUCAGUCGUGAGCCGACAACGACAACGCAACGGCGGC ((..(((.((((...))))...)))((..((...........((((......((((((((((((..........))))))))))))....---.)))).(((((.........)))))...))....)))). ( -28.90, z-score = -2.37, R) >droEre2.scaffold_4784 19466274 119 - 25762168 CCAAGCUCAAAGAUUCUUUAUAAGCGCAGGCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUGUAUGCAAAUAUCCCC---AUGUCAGUCGUGAGCCGACAACGACAACG---------- ((..(((.((((...))))...)))...))......(((...((((......((((((((((((..........))))))))))))....---.)))).((((.....))))...)))....---------- ( -25.40, z-score = -3.17, R) >droAna3.scaffold_13337 6069020 128 + 23293914 CCAAGCUCAAAGAUUCUUUAUAAGCGCAGGCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUAUCUCC---AUGUCAGUCGUGAGCCGACAACGACAACACACAGCCGC- ....(((.((((...))))...)))((.(((...........((((......((((((((((((..........))))))))))))....---.)))).(((((.........)))))........)))))- ( -28.70, z-score = -3.87, R) >droWil1.scaffold_180949 4801766 106 - 6375548 CAAAGCUCAAAGAUUCUUUAUAAGCGC--GCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUAUCCUCCUGAUGUCAGUCGUGAGG------------------------ ....(((.((((...))))...)))..--.......(((...))).......((((((((((((..........))))))))))))((((.((((....)))).))))------------------------ ( -20.10, z-score = -1.99, R) >droVir3.scaffold_13049 13253183 116 - 25233164 -CAAGCUCAAAGAUUCUUUAUAAGCGC--GCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUACCCC----GUGUCAGUCGUGAGCCG-CCGUCGUCGUC-GC------- -...(((((..((((........((((--(......(((...)))........(((((((((((..........)))))))))))...)----)))).)))).)))))((-.((....)).)-).------- ( -22.70, z-score = -1.63, R) >droGri2.scaffold_15110 18693088 123 - 24565398 -CAAGCUCAAAGAUUCUUUAUAAGCGC--GCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUAACCC----GUGUCAGUCGUGAGCCGCCCGUCGUCGCA-ACGGCUAC- -...(((((..((((........((((--(......(((...)))........(((((((((((..........)))))))))))...)----)))).)))).))))).....(((((....-)))))...- ( -26.70, z-score = -2.01, R) >consensus CCAAGCUCAAAGAUUCUUUAUAAGCGCAGGCACAAAGUCAAAGACAAUCAAAAUAUUUGCAUAUUACCAAUUUUAUAUGCAAAUAUCCCC___AUGUCAGUCGUGAGCCGACAACGACAACG_AA_G_CG__ ....(((((..((((........((....))...........((((......((((((((((((..........))))))))))))........)))))))).)))))........................ (-22.39 = -22.65 + 0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:15 2011