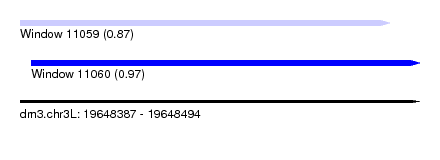
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,648,387 – 19,648,494 |
| Length | 107 |
| Max. P | 0.967630 |

| Location | 19,648,387 – 19,648,486 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.89 |
| Shannon entropy | 0.36239 |
| G+C content | 0.39793 |
| Mean single sequence MFE | -21.39 |
| Consensus MFE | -14.31 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19648387 99 + 24543557 ---------CAGAAGUAAUAGCAAUAA-UAACGCUCGUCAUAACAAUGGCAAACAAUGCAAUCAAAGCGAUUCGCAACACUUGCGCAUCCUUUGAGCCAACAAAUCGCA- ---------...........((.....-........(((((....))))).......((..((((((.(((.((((.....)))).))))))))))).........)).- ( -20.60, z-score = -1.49, R) >droSim1.chr3L 18974501 99 + 22553184 ---------CAGAAGUAAUAGCAAUAA-UAACGCUCGUCAUAACAAUGGCAAACAAUGCAAUCAAAGCGAUUCGCAACACUUGCGCAUCCUUUGAGCCAACAAAUCGCA- ---------...........((.....-........(((((....))))).......((..((((((.(((.((((.....)))).))))))))))).........)).- ( -20.60, z-score = -1.49, R) >droSec1.super_29 351241 99 + 700605 ---------CAGAAGUAAUAGCAAUAA-UAACGCUCGUCAUAACAAUGGCAAACAAUGCAAUCAAAGCGAUUCGCAACACUUGCGCAUCCUUUGAGCCAACAAAUCGCA- ---------...........((.....-........(((((....))))).......((..((((((.(((.((((.....)))).))))))))))).........)).- ( -20.60, z-score = -1.49, R) >droYak2.chr3L 3301726 105 - 24197627 ---CAGAAGCAAUAGCAAUAGUAAUAA-UAACGCUCGUCAUAACAAAGGCAAACAAUGCAAUCAAAGCGAUUCGCAACACUUGCGCAUCCUUUGAGCGACCAAAUCGCA- ---.....(((..(((...........-....))).(((........)))......)))..((((((.(((.((((.....)))).)))))))))((((.....)))).- ( -23.26, z-score = -1.89, R) >droEre2.scaffold_4784 19447551 105 + 25762168 ---CAGAAGUAAUAGUAUUAGUAAUAA-UACCGCUCGUCAUAACAAUGGCAAACAAUGCAAUCAAAGCGAUUCGCAACACUUGCGCAUCCUUUGAGCGACCGAAUCGCA- ---.....(((...((((((....)))-))).....(((((....)))))......)))..((((((.(((.((((.....)))).)))))))))((((.....)))).- ( -28.40, z-score = -3.18, R) >droAna3.scaffold_13337 6049899 99 - 23293914 ---------GGGAAAUAAUAAUAAUAA-UAACUCUCGUCAUAACAAUGGCAAACAAUGCAAUCAAAGCGAUUGGCAACACUUGCGCAUCCUUUGAGCGAACAAAUCGUA- ---------((((..............-........(((((....)))))................((((.((....)).))))...))))....((((.....)))).- ( -17.70, z-score = -1.04, R) >dp4.chrXR_group8 7721164 103 + 9212921 GUCCCAGGGAUAUACUUGUAAUAAUAA-UAAUUCUCGUCAUAACAAUAGCAAACAAUGCAAUCAAAGCGAUUUGCAACACUUGCGCAUCCUUUGAGCGAGGAAA------ ((..((((......))))..)).....-...(((((((..........(((.....)))..((((((.(((.((((.....)))).))))))))))))))))..------ ( -25.10, z-score = -2.73, R) >droVir3.scaffold_13049 13229087 97 + 25233164 -------------AACAAUAAUAAUAAGUAAUGCUCACAAUAACAAUACCAAACAAUGCAAUCAAAGCGAUUUGCAACACUUGCGCAUCCUUUGAAGGAAGCCAAUACGA -------------..............(((..(((..(...........((.....))...((((((.(((.((((.....)))).))))))))).)..)))...))).. ( -14.90, z-score = -1.20, R) >consensus _________CAGAAGUAAUAGUAAUAA_UAACGCUCGUCAUAACAAUGGCAAACAAUGCAAUCAAAGCGAUUCGCAACACUUGCGCAUCCUUUGAGCGAACAAAUCGCA_ ....................................(((((....)))))...........((((((.(((.((((.....)))).)))))))))............... (-14.31 = -14.90 + 0.59)


| Location | 19,648,390 – 19,648,494 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.79 |
| Shannon entropy | 0.34699 |
| G+C content | 0.38025 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -14.31 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19648390 104 + 24543557 ------AAGUAAUAGCAAUAA-UAACGCUCGUCAUAACAAUGGCAAACAAUGCAAUCAAAGCGAUUCGCAACACUUGCGCAUCCUUUGAGCCAACAAAUCGCAGAGUAAAC ------...............-....(((((((((....)))))......(((..((((((.(((.((((.....)))).)))))))))(....).....))))))).... ( -23.30, z-score = -2.31, R) >droSim1.chr3L 18974504 104 + 22553184 ------AAGUAAUAGCAAUAA-UAACGCUCGUCAUAACAAUGGCAAACAAUGCAAUCAAAGCGAUUCGCAACACUUGCGCAUCCUUUGAGCCAACAAAUCGCAGAGUAAAC ------...............-....(((((((((....)))))......(((..((((((.(((.((((.....)))).)))))))))(....).....))))))).... ( -23.30, z-score = -2.31, R) >droSec1.super_29 351244 104 + 700605 ------AAGUAAUAGCAAUAA-UAACGCUCGUCAUAACAAUGGCAAACAAUGCAAUCAAAGCGAUUCGCAACACUUGCGCAUCCUUUGAGCCAACAAAUCGCAGAGUAAAC ------...............-....(((((((((....)))))......(((..((((((.(((.((((.....)))).)))))))))(....).....))))))).... ( -23.30, z-score = -2.31, R) >droYak2.chr3L 3301729 110 - 24197627 AAGCAAUAGCAAUAGUAAUAA-UAACGCUCGUCAUAACAAAGGCAAACAAUGCAAUCAAAGCGAUUCGCAACACUUGCGCAUCCUUUGAGCGACCAAAUCGCAGAGUAAAC ..((....))...........-....(((((((........)))...........((((((.(((.((((.....)))).)))))))))((((.....)))).)))).... ( -25.60, z-score = -2.57, R) >droEre2.scaffold_4784 19447554 110 + 25762168 AAGUAAUAGUAUUAGUAAUAA-UACCGCUCGUCAUAACAAUGGCAAACAAUGCAAUCAAAGCGAUUCGCAACACUUGCGCAUCCUUUGAGCGACCGAAUCGCAGAGCAAAC ........((((((....)))-))).(((((((((....)))))...........((((((.(((.((((.....)))).)))))))))((((.....)))).)))).... ( -33.80, z-score = -4.73, R) >droAna3.scaffold_13337 6049902 104 - 23293914 ------AAAUAAUAAUAAUAA-UAACUCUCGUCAUAACAAUGGCAAACAAUGCAAUCAAAGCGAUUGGCAACACUUGCGCAUCCUUUGAGCGAACAAAUCGUAAAAUAAAC ------...............-........(((((....)))))...........((((((.(((.((((.....))).))))))))))((((.....))))......... ( -16.60, z-score = -1.48, R) >dp4.chrXR_group8 7721171 103 + 9212921 -GGAUAUACUUGUAAUAAUAA-UAAUUCUCGUCAUAACAAUAGCAAACAAUGCAAUCAAAGCGAUUUGCAACACUUGCGCAUCCUUUGAGCGAGGAAAAAAAAAA------ -....................-...(((((((..........(((.....)))..((((((.(((.((((.....)))).)))))))))))))))).........------ ( -22.10, z-score = -2.74, R) >droVir3.scaffold_13049 13229087 104 + 25233164 -------AACAAUAAUAAUAAGUAAUGCUCACAAUAACAAUACCAAACAAUGCAAUCAAAGCGAUUUGCAACACUUGCGCAUCCUUUGAAGGAAGCCAAUACGACAAAAAA -------..............(((..(((..(...........((.....))...((((((.(((.((((.....)))).))))))))).)..)))...)))......... ( -14.90, z-score = -1.28, R) >consensus ______AAGUAAUAGUAAUAA_UAACGCUCGUCAUAACAAUGGCAAACAAUGCAAUCAAAGCGAUUCGCAACACUUGCGCAUCCUUUGAGCGAACAAAUCGCAGAGUAAAC ..............................(((((....)))))...........((((((.(((.((((.....)))).)))))))))...................... (-14.31 = -14.90 + 0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:12 2011