| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,608,499 – 19,608,602 |
| Length | 103 |
| Max. P | 0.568098 |

| Location | 19,608,499 – 19,608,602 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 69.06 |
| Shannon entropy | 0.68132 |
| G+C content | 0.51398 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -10.56 |
| Energy contribution | -9.34 |
| Covariance contribution | -1.23 |
| Combinations/Pair | 1.77 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.568098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
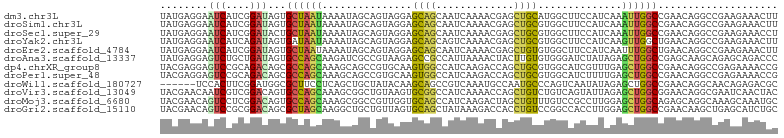
>dm3.chr3L 19608499 103 - 24543557 UAUGAGGAAUCAUCGGAUAGUGCUAAUAAAAUAGCAGUAGGAGCAGCAAUCAAAACGAGCUGCAUGGCUUCCAUCAAAUUGGCCGAACAGGCCGAAGAAACUU ..(((((((((((....((.(((((......))))).))...(((((..((.....))))))))))).)))).)))..((((((.....))))))........ ( -31.50, z-score = -3.23, R) >droSim1.chr3L 18935195 103 - 22553184 UAUGAGGAAUCAUCGGAUAGUGCUAAUAAAAUAGCAGUAGGAGCAGCAAUCAAAACGAGCUGCGUGGCUUCCAUCAAAUUGGCCGAACAGGCCGAAGAAACUU ..(((((((((((....((.(((((......))))).))...(((((..((.....))))))))))).)))).)))..((((((.....))))))........ ( -31.80, z-score = -3.05, R) >droSec1.super_29 312057 103 - 700605 UAUGAGGAAUCAUCGGAUACUGCUAAUAAAAUAGCAGUAGGAGCAGCAAUCAAAACGAGCUGCGUGGCUUCCAUCAAAUUGGCCGAACAGGCCGAAGAAACCU ..(((((((((((....((((((((......))))))))...(((((..((.....))))))))))).)))).)))..((((((.....))))))........ ( -35.70, z-score = -4.19, R) >droYak2.chr3L 3262371 103 + 24197627 UAUGAGGAAUCAUCAGAUAGUGAUAAUAAAAUAGCAGUAGGAGCAGCAGUCAAAACGAGCUGCGUGGCUUCCAUCAAGUUGGCUGAACAGGCCGAAGAAACUU ..(((((((((((......)))))...........(((....(((((.((....))..)))))...)))))).)))..((((((.....))))))........ ( -24.60, z-score = -0.90, R) >droEre2.scaffold_4784 19407358 103 - 25762168 UAUGAGGAAUCAUCGGAUAGUGCUAAUAAAAUAGCAGUAGGAGCAGCAAUCAAAACGAGCUGUGUGGCUUCCAUCAAUUUGGCUGAACAGGCCGAAGAAACUU ..(((((((((((....((.(((((......))))).))...(((((..((.....))))))))))).)))).))).(((((((.....)))))))....... ( -27.70, z-score = -1.68, R) >droAna3.scaffold_13337 6012108 103 + 23293914 UAUGAGGAGUCUGCUGAUAGUGCCAGCAAGAUCGCCGUAAGAGCCGCCAUUAAAACUACUUGUGUGGGAUCUAUAGAGCUGGCCGAGCAAGCAGAGCAGACCC .....((.(((((((....(.((((((.((((((((....).))...........((((....))))))))).....)))))))..(....)..))))))))) ( -34.20, z-score = -2.02, R) >dp4.chrXR_group8 7684231 103 - 9212921 UACGAGGAGUCCGCAGACAGCGCCAGCAAAGCAGCCGUGCAAGUGGCCAUCAAGACCAGCUGCGUGGCAUCGUUUGAGCUGGCCGAACAGGCCGAGAAAACCG .....(((((...(((((.((....))......((((((((..(((.(.....).)))..))))))))...))))).)))((((.....)))).......)). ( -33.50, z-score = -0.22, R) >droPer1.super_48 462675 103 - 581423 UACGAGGAGUCCGCAGACAGCGCCAGCAAAGCAGCCGUGCAAGUGGCCAUCAAGACCAGCUGCGUGGCAUCUUUUGAGCUGGCCGAACAGGCCGAGAAAACCG .....((..(((((.....(.((((((......((((((((..(((.(.....).)))..)))))))).((....)))))))))......).)).))...)). ( -31.40, z-score = 0.33, R) >droWil1.scaffold_180727 1877148 97 + 2741493 ------UCCACUUCGGAUGGCGCUUCCUCAGCUGCUAUACAAGCAGCCGUCAAAUGCCAAUGCCCAGUCAAUAUAGAGCUGGCCGAACAGGCAACAGAGACGC ------....((..((((((((((.....)))((((.....))))))))))...((((..((.((((((......)).)))).))....)))).)..)).... ( -27.70, z-score = -0.64, R) >droVir3.scaffold_13049 13189907 103 - 25233164 UACGAACAAUCGUCGGACAGUGCCAGCAAAGCGGCUGUAAGUGCGGCCAUCAAAACCAGCUGUCUGUCAGUAUUAGAGCUGGCGGAACAGGCGAAUCAACUAC .........((((((((((((((.......))((((((....))))))..........)))))))((((((......))))))......)))))......... ( -33.80, z-score = -1.81, R) >droMoj3.scaffold_6680 10591515 103 + 24764193 UACGAACAGUCCUCGGACAGUGCCAGCAAAGCGGCCGUUGGUGCAGCCAUCAAGACUAGCUGUUUGUCCGCCUUGGAGCUGGCAGAGCAGGCAAAGCAAAUGC ........(((....)))..(((((((.(((((((..((((((....)))))).....))))))).(((.....))))))))))..(((.((...))...))) ( -32.00, z-score = 0.78, R) >droGri2.scaffold_15110 18628944 103 - 24565398 UACGAACAGUCCGCGGACAGCGCUAGCAAGGCUGCUGUUAGUGCAGCUAUAAAGACCACCUGUCCGGCCACCUUGGAGCUGGCCGAACAAGCUGAGCAUCUGC ............(((((..((..((((..((((((.......))))))............(((.((((((.(.....).)))))).))).)))).)).))))) ( -35.30, z-score = -0.42, R) >consensus UAUGAGGAAUCAUCGGAUAGUGCUAGCAAAACAGCAGUAGGAGCAGCCAUCAAAACCAGCUGCGUGGCUUCCAUCGAGCUGGCCGAACAGGCCGAAGAAACCC ........(((....)))...((((((...............((((.............))))..............)))))).................... (-10.56 = -9.34 + -1.23)

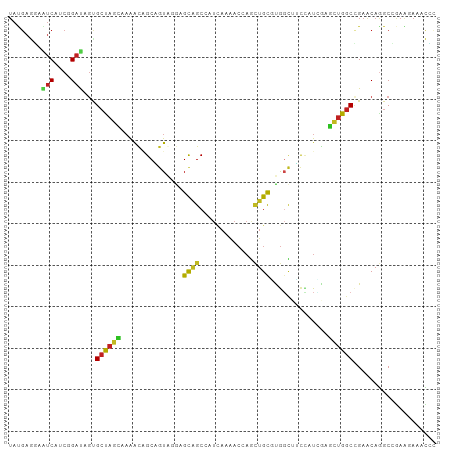
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:10 2011