| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,736,172 – 5,736,290 |
| Length | 118 |
| Max. P | 0.879036 |

| Location | 5,736,172 – 5,736,290 |
|---|---|
| Length | 118 |
| Sequences | 14 |
| Columns | 131 |
| Reading direction | reverse |
| Mean pairwise identity | 72.52 |
| Shannon entropy | 0.59200 |
| G+C content | 0.42496 |
| Mean single sequence MFE | -29.54 |
| Consensus MFE | -16.34 |
| Energy contribution | -16.10 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
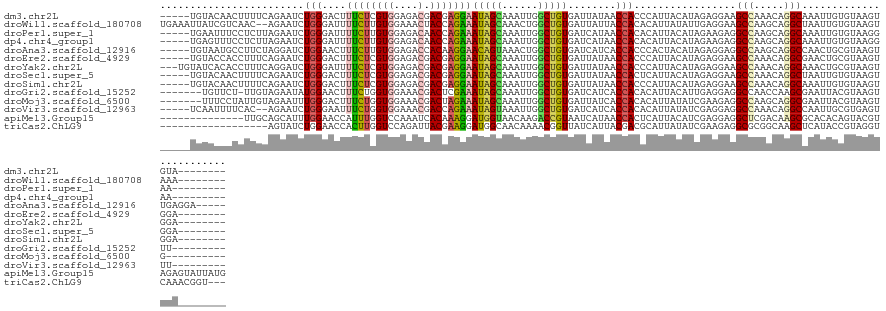
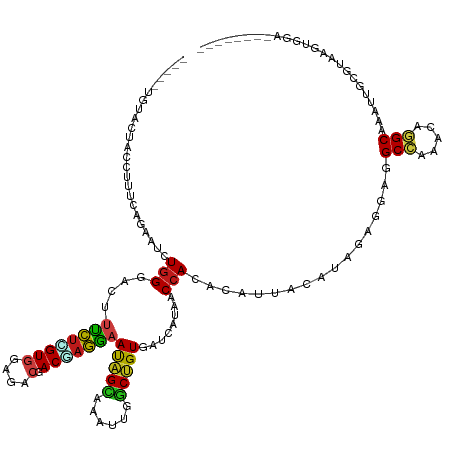
>dm3.chr2L 5736172 118 - 23011544 -----UGUACAACUUUUCAGAAUCUGGGACUUUCUCGUGGAGACGACGAGGAAUAGCAAAUUGGCUGUGAUUAUAACCACCCAUUACAUAGAGGAAGCCAAACAGGCAAAUUGUGUAAGUGUA-------- -----..((((.............((((...((((((((....).)))))))(((((......)))))...........))))((((((((.....(((.....)))...)))))))).))))-------- ( -30.70, z-score = -1.43, R) >droWil1.scaffold_180708 4111809 121 + 12563649 UGAAAUUAUCGUCAAC--AGAAUCUGGGAUUUUCUUGUGGAAACUACCAGAAAUAGCAAACUGGCUGUGAUUAUUACCACACAUUAUAUUGAGGAAGCCAAGCAGGCUAAUUGUGUAAGUAAA-------- ..............((--(.(((((((..(((((....)))))...)))))..((((....((((((((........))).((......))....))))).....)))).)).))).......-------- ( -24.50, z-score = -0.15, R) >droPer1.super_1 9615113 117 + 10282868 -----UGAAUUUCCUCUUAGAAUCUGGGAUUUUCUUGUGGAGACAACCAGAAAUAGCAAAUUGGCUGUGAUCAUAACCACACAUUACAUAGAAGAGGCCAAGCAGGCAAAUUGUGUAAGGAA--------- -----.....((((.....((.(((((..(((((....)))))...))))).(((((......)))))..))...........((((((((.....(((.....)))...))))))))))))--------- ( -27.30, z-score = -0.68, R) >dp4.chr4_group1 4607447 117 + 5278887 -----UGAGUUUCCUCUUAGAAUCUGGGAUUUUCUUGUGGAGACAACCAGAAAUAGCAAAUUGGCUGUGAUCAUAACCACACAUUACAUAGAAGAGGCCAAGCAGGCAAAUUGUGUAAGGAA--------- -----...((((((.(..((((((...))))))...).))))))..((.((.(((((......)))))..))...........((((((((.....(((.....)))...))))))))))..--------- ( -28.00, z-score = -0.59, R) >droAna3.scaffold_12916 11176998 121 - 16180835 -----UGUAAUGCCUUCUAGGAUCUGGAACUUUCUUGUGGAGACCACAAGGAACAGUAAACUGGCUGUGAUCAUCACCACCCACUACAUAGAGGAGGCCAAGCAGGCCAACUGCGUAAGUUGAGGA----- -----....((((((((((.....(((....(((((((((...)))))))))(((((......)))))............))).....)))))).((((.....))))....))))..........----- ( -34.90, z-score = -0.29, R) >droEre2.scaffold_4929 5826176 118 - 26641161 -----UGUACCACCUUUCAGAAUCUGGGACUUUCUCGUGGAGACGACGAGGAAUAGCAAAUUGGCUGUGAUUAUAACCACCCAUUACAUAGAGGAAGCCAAACAGGCGAACUGCGUAAGUGGA-------- -----.......(((((.......((((...((((((((....).)))))))(((((......)))))...........))))......)))))..(((.....)))...(..(....)..).-------- ( -33.92, z-score = -1.87, R) >droYak2.chr2L 8868580 120 + 22324452 ---UGUAUCACACCUUUCAGGAUCUGGGAUUUUCUCGUGGAGACGACGAGGAAUAGCAAAUUGGCUGUGAUUAUAACCACCCAUUACAUAGAGGAAGCCAAACAGGCAAACUGCGUAAGUGGA-------- ---.........(((((..(....((((...((((((((....).)))))))(((((......)))))...........))))...)..)))))..(((.....)))...(..(....)..).-------- ( -34.00, z-score = -1.75, R) >droSec1.super_5 3814881 118 - 5866729 -----UGUACAACUUUUCAGAAUCUGGGACUUUCUCGUGGAGACGACGAGGAAUAGCAAAUUGGCUGUGAUUAUAACCACUCAUUACAUAGAGGAAGCCAAACAGGCUAAUUGUGUAAGUGGA-------- -----.........(..(((...)))..)..((((((((....).)))))))(((((......)))))........(((((...(((((((....((((.....))))..)))))))))))).-------- ( -33.10, z-score = -2.16, R) >droSim1.chr2L 5532663 118 - 22036055 -----UGUACAACUUUUCAGAAUCUGGGACUUUCUCGUGGAGACGACGAGGAAUAGCAAAUUGGCUGUGAUUAUAACCACCCAUUACAUAGAGGAAGCCAAACAGGCAAAUUGUGUAAGUGGA-------- -----.........(..(((...)))..)..((((((((....).)))))))(((((......)))))........((((...((((((((.....(((.....)))...)))))))))))).-------- ( -30.70, z-score = -1.39, R) >droGri2.scaffold_15252 3282589 114 - 17193109 -------UGUUCU-UUGUAGAAUAUGGAACUUUCUGGUGGAAACGACUCGAAAUAGCAAAUUGGCUGUGAUCAUCACCACACAUUACAUUGAGGAGGCCAACCAAGCGAAUUACGUAAGUUU--------- -------((((((-....))))))..((((((..((((((...(..(((((...(((......)))(((........)))........)))))..).)).)))).(((.....)))))))))--------- ( -24.30, z-score = -0.12, R) >droMoj3.scaffold_6500 14232654 114 - 32352404 -------UUUCCUAUUGUAGAAUUUGGGACUUUCUGGUGGAAACGACUAGAAAUAGCAAAUUGGCUGUGAUUAUCACCACACAUUAUAUCGAAGAGGCCAAGCAGGCGAAUUACGUAAGUG---------- -------..(((((..........))))).(((((((((....).))))))))..((...((((((.((((................))))....))))))....))....(((....)))---------- ( -27.09, z-score = -1.15, R) >droVir3.scaffold_12963 3777887 115 - 20206255 -----UCAAUUUUCAC--AGAAUCUGGGAAUUUCUGGUGGAAACGACCAGAAAUAGUAAAUUGGCUGUGAUCAUCACCACACAUUAUAUCGAGGAGGCCAAACAGGCCAAUUGCGUGAGUUU--------- -----......(((((--....(((..((((((((((((....).))))))))).((((..((...(((.....)))..))..)))).))..)))((((.....))))......)))))...--------- ( -31.90, z-score = -1.95, R) >apiMel3.Group15 4127243 117 + 7856270 --------------UUGCAGCAUUUGGAACCAUUUGGUCCAAAUCACAAAGGAUGGUAACAAGACCGUAAUCAUAACCACUCAUUACAUCGAGGAGGCUCGACAAGCGCACACAGUACGUAGAGUAUUAUG --------------.(((.(((((((((.((....)))))))))......(((((((.((......)).)))))..))..........(((((....)))))...)))))((.(((((.....))))).)) ( -26.10, z-score = -1.31, R) >triCas2.ChLG9 12395133 110 - 15222296 ------------------AGUAUCUGGAACCACUUGGUCCAGAUUACGAAGGAUGGCAACAAAACGGUUAUCAUUACGACGCAUUAUAUCGAAGAGGCGCGGCAAGCUCAUACCGUAGGUCAAACGGU--- ------------------.(((((((((.((....)))))))).)))((..((((..(((......)))..)))).((.(((.((........)).)))))......))..(((((.......)))))--- ( -27.00, z-score = -0.69, R) >consensus _____UGUACUACCUUUCAGAAUCUGGGACUUUCUCGUGGAGACGACGAGGAAUAGCAAAUUGGCUGUGAUCAUAACCACACAUUACAUAGAGGAGGCCAAACAGGCAAAUUGCGUAAGUGGA________ ........................(((....((((((((....).)))))))(((((......)))))........))).................(((.....)))........................ (-16.34 = -16.10 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:46 2011