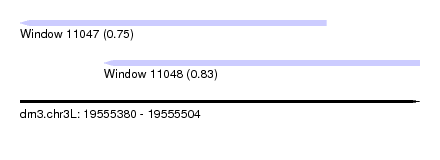
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,555,380 – 19,555,504 |
| Length | 124 |
| Max. P | 0.826358 |

| Location | 19,555,380 – 19,555,475 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 96.42 |
| Shannon entropy | 0.06342 |
| G+C content | 0.55698 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -32.38 |
| Energy contribution | -33.22 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.746148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
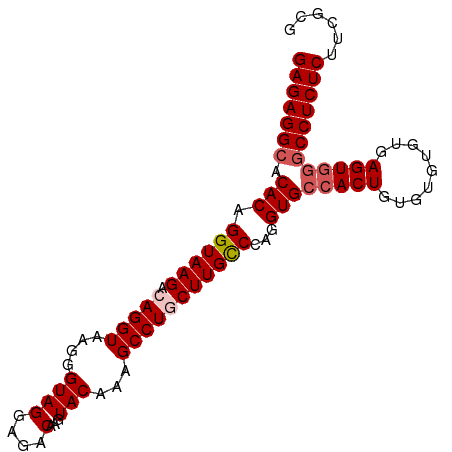
>dm3.chr3L 19555380 95 - 24543557 GAGAGGCACACAGGUAAGACAGGUAAGGGUAGGAGACAAGUACAAAGCCUGCUUGCCCAGGUGCCACUGUAUGUGUGAGUGUACCUCUCUUCGCG ((((((.((((.((((((.(((((....((((....)...)))...)))))))))))((.((((....)))).))...)))).))))))...... ( -37.40, z-score = -2.51, R) >droSim1.chr3L 18887591 95 - 22553184 GAGAGGCACACAGGUAAGACAGGUAAGGGUAGGAGACAAGUACAAAGCCUGCUUGCCCAGGUGCCACUGUGUGCGUGAGUGGGCCUCUCUUCGCG (((((((.(((.((((((.(((((....((((....)...)))...)))))))))))((.(..(......)..).)).))).)))))))...... ( -40.60, z-score = -2.55, R) >droSec1.super_29 258575 95 - 700605 GAGAGGCACACAGGUAAGACAGGUAAGGGUAGGAGACAAGUACAAAGCCUGCUUGCCCAGGUGCCACUGUGUGUGUGAGUUGGCCUCUCUUCGCG ((((((((((((((((((.(((((....((((....)...)))...)))))))))))(((......)))..)))))......)))))))...... ( -36.30, z-score = -1.55, R) >droYak2.chr3L 3205377 95 + 24197627 GAGAGGCACACAGGUAAGACAGGUAAGGGUAGGAGACAAGUACAAAGCCUACUUGUCCAGGUGCCACUGUGUGUGUGAGUGGGCCUCUCUUCGCG (((((((.(((.....(.((((.....((((.(.((((((((.......)))))))))...)))).)))).)......))).)))))))...... ( -34.60, z-score = -1.35, R) >droEre2.scaffold_4784 19352922 94 - 25762168 GAGAGGCACACAGGUAAGACAGGUAAGGGUAGGAGACAAGUACAAAGCCUACUUGCCC-GGUGCCACUGUGUGUGUGAGUGGGCCUCUCUUCGCG (((((((.(((.((((((..((((....((((....)...)))...)))).)))))).-.)))(((((.........))))))))))))...... ( -35.10, z-score = -1.32, R) >consensus GAGAGGCACACAGGUAAGACAGGUAAGGGUAGGAGACAAGUACAAAGCCUGCUUGCCCAGGUGCCACUGUGUGUGUGAGUGGGCCUCUCUUCGCG (((((((.(((.((((((.(((((....((((....)...)))...)))))))))))...)))(((((.........))))))))))))...... (-32.38 = -33.22 + 0.84)

| Location | 19,555,406 – 19,555,504 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 98.16 |
| Shannon entropy | 0.03201 |
| G+C content | 0.55624 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -30.18 |
| Energy contribution | -30.62 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
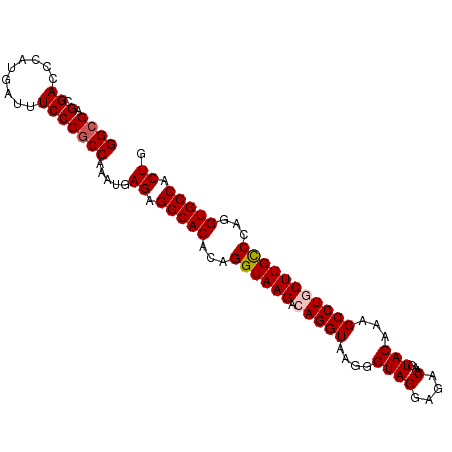
>dm3.chr3L 19555406 98 - 24543557 GGGGAGCGACCCAUGAUUUCCCGCCAAAUGAGAGGCACACAGGUAAGACAGGUAAGGGUAGGAGACAAGUACAAAGCCUGCUUGCCCAGGUGCCACUG ((((((((.....)).))))))........((.(((((...((((((.(((((....((((....)...)))...)))))))))))...))))).)). ( -35.10, z-score = -2.10, R) >droSim1.chr3L 18887617 98 - 22553184 GGCGAGCGACCCAUGAUUUCCCGCCAAAUGAGAGGCACACAGGUAAGACAGGUAAGGGUAGGAGACAAGUACAAAGCCUGCUUGCCCAGGUGCCACUG ((((.(.((.........))))))).....((.(((((...((((((.(((((....((((....)...)))...)))))))))))...))))).)). ( -35.10, z-score = -2.53, R) >droSec1.super_29 258601 98 - 700605 GGCGAGCGACCCAUGAUUUCCCGCCAAAUGAGAGGCACACAGGUAAGACAGGUAAGGGUAGGAGACAAGUACAAAGCCUGCUUGCCCAGGUGCCACUG ((((.(.((.........))))))).....((.(((((...((((((.(((((....((((....)...)))...)))))))))))...))))).)). ( -35.10, z-score = -2.53, R) >droYak2.chr3L 3205403 98 + 24197627 GGCGAGCGACCCAUGAUUUCCCGCCAAAUGAGAGGCACACAGGUAAGACAGGUAAGGGUAGGAGACAAGUACAAAGCCUACUUGUCCAGGUGCCACUG ((((.(.((.........))))))).....((.(((((...((((((..((((....((((....)...)))...)))).)))).))..))))).)). ( -26.30, z-score = -0.26, R) >droEre2.scaffold_4784 19352948 97 - 25762168 GGCGAGCGACCCAUGAUUUCCCGCCAAAUGAGAGGCACACAGGUAAGACAGGUAAGGGUAGGAGACAAGUACAAAGCCUACUUGCCC-GGUGCCACUG ((((.(.((.........))))))).....((.(((((.(.((((((..((((....((((....)...)))...)))).)))))).-)))))).)). ( -30.40, z-score = -1.50, R) >consensus GGCGAGCGACCCAUGAUUUCCCGCCAAAUGAGAGGCACACAGGUAAGACAGGUAAGGGUAGGAGACAAGUACAAAGCCUGCUUGCCCAGGUGCCACUG ((((.(.((.........))))))).....((.(((((...((((((.(((((....((((....)...)))...)))))))))))...))))).)). (-30.18 = -30.62 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:38:02 2011