| Sequence ID | dm3.chr3L |
|---|---|
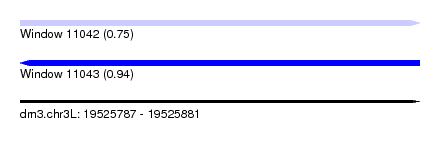
| Location | 19,525,787 – 19,525,881 |
| Length | 94 |
| Max. P | 0.938294 |

| Location | 19,525,787 – 19,525,881 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 86.83 |
| Shannon entropy | 0.27585 |
| G+C content | 0.58380 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -26.00 |
| Energy contribution | -26.69 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.747277 |
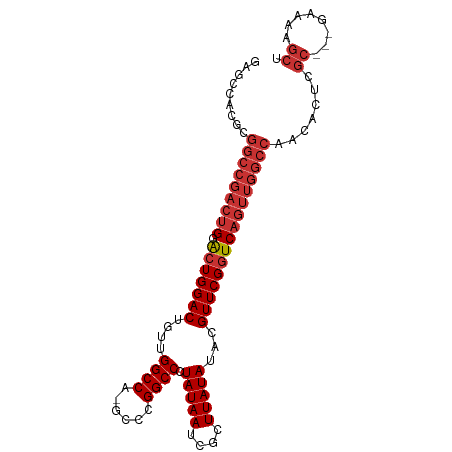
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19525787 94 + 24543557 AGCUUUUC---GCGACUGUUGGCCAACUGACCGAACGUAUAUAAGCGAUUAUAGGGCCGGGC-UGGCCAACAGUCCAGUCCAGUCGGCCGCGUGGCUC ((((...(---((((((((((((((.....(((..(.((((........)))).)..)))..-))))))))))))..(.((....)).)))).)))). ( -39.70, z-score = -2.59, R) >droSec1.super_29 228505 94 + 700605 AGCUUUUC---GCGACUGUUGGCCAACUGACCGAACGUAUAUAAGCGAUUAUAGGGCCGGGC-UGGCCAACAGUCCAGUCCAGUCGGCCGCGUGGCUC ((((...(---((((((((((((((.....(((..(.((((........)))).)..)))..-))))))))))))..(.((....)).)))).)))). ( -39.70, z-score = -2.59, R) >droYak2.chr3L 3181394 94 - 24197627 AGCUUUUC---GCGAGUGUUGGCCAACUGACCGAACGUAUAUAAGCGAUUAUAGGGCCGGGC-UGGCCAACAGUCCAGUCCAGUCGGCCGCGUGGCUC ((((...(---((((.(((((((((.....(((..(.((((........)))).)..)))..-))))))))).))..(.((....)).)))).)))). ( -35.80, z-score = -1.38, R) >droEre2.scaffold_4784 19323555 93 + 25762168 AGCUUUUC---GCG-GUGUUGGCCAACUGACCGAACGUAUAUAAGCGAUUAUAGGGCCGGGC-UGGCCAACAGUCCAGUCCAGUCGGCCGCGUGGCUC ((((...(---(((-((((..(....)..))(((.(((......)))......((((.((((-((.....)))))).))))..))))))))).)))). ( -38.20, z-score = -1.89, R) >dp4.chrXR_group8 7601529 97 + 9212921 AGCUUUUCUGUGCGAGUGUUGGCCAACUGACCGAACGUAUAUAAGCGAUUAUAGGGCCGGGC-UGGCCAACAGUCCAGUCCAGUCGGCCGCGCUGCCU ...........((.(((((.((((.(((.......(((......)))......((((.((((-((.....)))))).))))))).))))))))))).. ( -36.40, z-score = -1.12, R) >droPer1.super_48 379598 97 + 581423 AGCUUUUCUGUGCGAGUGUUGGCCAACUGACCGAACGUAUAUAAGCGAUUAUAGGGCCGGGC-UGGCCAACAGUCCAGUCCAGUCGGCCGCGCUGCCU ...........((.(((((.((((.(((.......(((......)))......((((.((((-((.....)))))).))))))).))))))))))).. ( -36.40, z-score = -1.12, R) >droSim1.chr3L 18856427 94 + 22553184 AGCUUUUC---GCGACUGUUGGCCAACUGACCGAACGUAUAUAAGCGAUUAUAGGGCCGGGC-UGGCCAACAGUCCAGUCCAGUCGGCCGCGUAGCUC ((((...(---((((((((((((((.....(((..(.((((........)))).)..)))..-))))))))))))..(.((....)).)))).)))). ( -39.70, z-score = -3.05, R) >droWil1.scaffold_180727 806549 74 + 2741493 ---------GCACCCGACUUGGCCAACUGACCGAACGUAUAUAAGCGAUUAUAGGGCCGGGCCUGGCCAUCAGUCCACCGCGU--------------- ---------((....((((((((((.....(((..(.((((........)))).)..)))...))))))..))))....))..--------------- ( -23.70, z-score = -0.80, R) >consensus AGCUUUUC___GCGAGUGUUGGCCAACUGACCGAACGUAUAUAAGCGAUUAUAGGGCCGGGC_UGGCCAACAGUCCAGUCCAGUCGGCCGCGUGGCUC .............((.(((((((((.....(((..(.((((........)))).)..)))...))))))))).))..........(((......))). (-26.00 = -26.69 + 0.69)

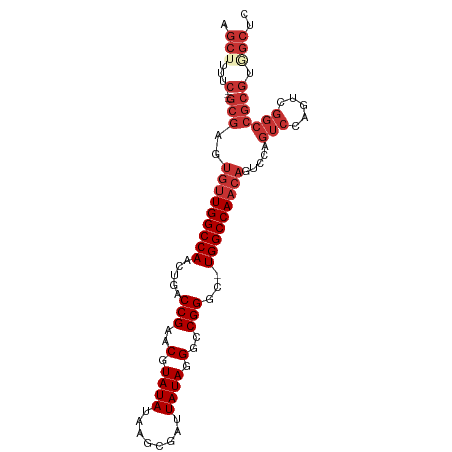
| Location | 19,525,787 – 19,525,881 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 86.83 |
| Shannon entropy | 0.27585 |
| G+C content | 0.58380 |
| Mean single sequence MFE | -35.71 |
| Consensus MFE | -29.33 |
| Energy contribution | -30.35 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19525787 94 - 24543557 GAGCCACGCGGCCGACUGGACUGGACUGUUGGCCA-GCCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAACAGUCGC---GAAAAGCU .(((..(((((((....)).)).((((((((((((-(((((..(.((((........)))).)..)))...))))))))))))))))---)....))) ( -38.80, z-score = -2.70, R) >droSec1.super_29 228505 94 - 700605 GAGCCACGCGGCCGACUGGACUGGACUGUUGGCCA-GCCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAACAGUCGC---GAAAAGCU .(((..(((((((....)).)).((((((((((((-(((((..(.((((........)))).)..)))...))))))))))))))))---)....))) ( -38.80, z-score = -2.70, R) >droYak2.chr3L 3181394 94 + 24197627 GAGCCACGCGGCCGACUGGACUGGACUGUUGGCCA-GCCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAACACUCGC---GAAAAGCU (((......(((((((((.(((((((....((((.-....)))).(((((....)))))...))))))))))))))))....)))((---.....)). ( -35.70, z-score = -2.08, R) >droEre2.scaffold_4784 19323555 93 - 25762168 GAGCCACGCGGCCGACUGGACUGGACUGUUGGCCA-GCCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAACAC-CGC---GAAAAGCU .(((..((((((((((((.(((((((....((((.-....)))).(((((....)))))...)))))))))))))))).....-.))---)....))) ( -35.30, z-score = -1.93, R) >dp4.chrXR_group8 7601529 97 - 9212921 AGGCAGCGCGGCCGACUGGACUGGACUGUUGGCCA-GCCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAACACUCGCACAGAAAAGCU ....(((..(((((((((.(((((((....((((.-....)))).(((((....)))))...)))))))))))))))).....((.....))...))) ( -35.50, z-score = -1.32, R) >droPer1.super_48 379598 97 - 581423 AGGCAGCGCGGCCGACUGGACUGGACUGUUGGCCA-GCCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAACACUCGCACAGAAAAGCU ....(((..(((((((((.(((((((....((((.-....)))).(((((....)))))...)))))))))))))))).....((.....))...))) ( -35.50, z-score = -1.32, R) >droSim1.chr3L 18856427 94 - 22553184 GAGCUACGCGGCCGACUGGACUGGACUGUUGGCCA-GCCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAACAGUCGC---GAAAAGCU .((((.(((((((....)).)).((((((((((((-(((((..(.((((........)))).)..)))...))))))))))))))))---)...)))) ( -40.50, z-score = -3.31, R) >droWil1.scaffold_180727 806549 74 - 2741493 ---------------ACGCGGUGGACUGAUGGCCAGGCCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAAGUCGGGUGC--------- ---------------..(((.(.((((..((((((((((..((..(((((....)))))...))..))))...)))))))))).).)))--------- ( -25.60, z-score = -0.08, R) >consensus GAGCCACGCGGCCGACUGGACUGGACUGUUGGCCA_GCCCGGCCCUAUAAUCGCUUAUAUACGUUCGGUCAGUUGGCCAACACUCGC___GAAAAGCU .........(((((((((.(((((((....((((......)))).(((((....)))))...)))))))))))))))).................... (-29.33 = -30.35 + 1.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:57 2011