
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,514,504 – 19,514,638 |
| Length | 134 |
| Max. P | 0.986951 |

| Location | 19,514,504 – 19,514,638 |
|---|---|
| Length | 134 |
| Sequences | 10 |
| Columns | 139 |
| Reading direction | forward |
| Mean pairwise identity | 57.14 |
| Shannon entropy | 0.88445 |
| G+C content | 0.36982 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -10.82 |
| Energy contribution | -11.07 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
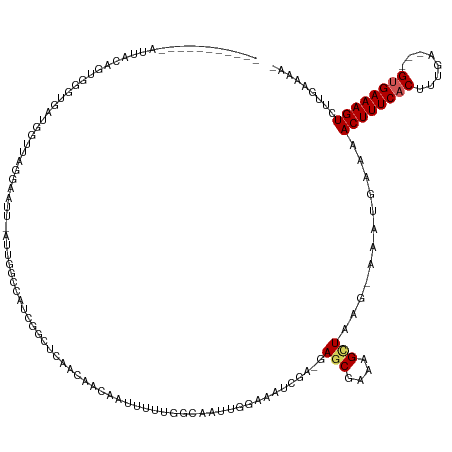
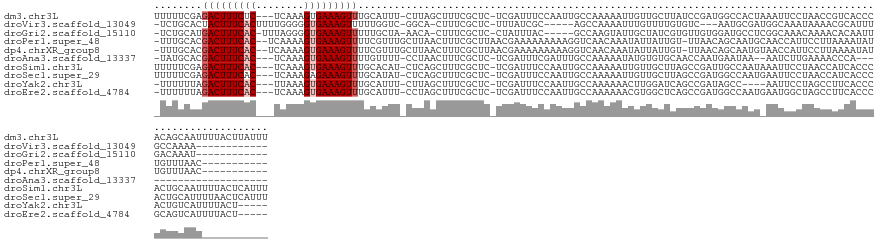
>dm3.chr3L 19514504 134 + 24543557 AAAUAAGUAAAAUUGCUGUGGGUGACGGUUAGGAAUUUAGUGGCCAUCGGAUAAGCAACAAUUUUUGGCAAUUGGAAAUCGA-GAGCGAAAGCUAAG-AAAUGCAAACUUUCACUUUGA---GAGAAAGUCUCGAAAAA .....((((....))))...(((((.((((.(.(.((((((.((((..((((........)))).)))).))))))..((..-.(((....)))..)-)..).).)))).)))))((((---((.....)))))).... ( -28.70, z-score = -0.44, R) >droVir3.scaffold_13049 13089386 115 + 25233164 ------------UUUUGGCAAAUGCGUUUUAUUUGCCAUCGCAUU---GACACAAAACAAAUUUUGGCU-----GCGAUAAA-GAGCGAAAG-UGCC-GACCAAAAACUUUCACCCCCAAAAGUGAAAGUAGUGCAGA- ------------...(((((((((.....)))))))))..(((((---.............(((((((.-----((......-..((....)-))).-).))))))((((((((........)))))))))))))...- ( -31.90, z-score = -2.66, R) >droGri2.scaffold_15110 23826059 117 + 24565398 ------------AUUUGUCAAUUGUGUUUUGUUUGCCGAGGCAUCCACAACACGAUAGCAAUACUUGGC-----GUAAAUAG-GAGCGAAAG-UGUU-UAGCAAAAACUUUCACCCCUAAA-GUGAAAGUCAUGCAGA- ------------...(((..((((((((.((..(((....)))..)).)))))))).)))((((((..(-----((......-..))).)))-))).-..(((...((((((((.......-))))))))..)))...- ( -33.80, z-score = -2.75, R) >droPer1.super_48 368972 124 + 581423 -----------GUUAAACAAUAUUUUAAGGAAUGGUUGCAUUGCUGUUAA-ACAAUAAUAUUUGUUGACCUUUUUUUUUCGUUAAGCGAAAGUUAAGCAAACGAAAACUUUCACUUUUGA--GUGAAAGUCGUGCAAA- -----------........................((((.((((((((((-.(((......))))))))......((((((.....))))))...)))))((((...((((((((....)--))))))))))))))).- ( -27.80, z-score = -1.57, R) >dp4.chrXR_group8 7590875 124 + 9212921 -----------GUUAAACAAUAUUUUAAGGAAUGGUUACAUUGCUGUUAA-ACAAUAAUAUUUGUUGACCUUUUUUUUUCGUUAAGCGAAAGUUAAGCAAACGAAAACUUUCACUUUUGA--GUGAAAGUCGUGCAAA- -----------(((.(((........((((((((((......)))))).(-((((......)))))..))))....(((((.....)))))))).)))..((((...((((((((....)--))))))))))).....- ( -26.60, z-score = -1.53, R) >droAna3.scaffold_13337 6434406 109 + 23293914 ----------------------UGGGUUUUCAAGAUU--UUAUUCAUUGGUUGCACACAUAUUUUUGGCAAAUCGAAAUCGA-GAGCGAAAGUUAGG-AAAACAAAACUUUCACUUUGA---GUGAAAGUCGUGCAUA- ----------------------.......((((((..--....)).)))).(((((.......((((.....(((....)))-.(((....)))...-....))))((((((((.....---)))))))).)))))..- ( -24.30, z-score = -1.22, R) >droSim1.chr3L 18844988 134 + 22553184 AAAUGAGUAAAAUUGCAGUGGGUGAUGGUUAGGAAUUUAUUGGCAAUCGGCUAAGCAACAAUUUUUGGCAAUUGGAAAUCGA-GAGCGAAAGCUGAG-AUGUGCAAACUUUCACUUUGA---GUGAAAGUCUCGAAAAA ..........((((((..(((.((((.(((((.......))))).)))).)))..(((......))))))))).....((((-((((....)))...-........((((((((.....---))))))))))))).... ( -34.30, z-score = -1.66, R) >droSec1.super_29 217596 134 + 700605 AAAUGAGUUAAAAUGCAGUGGGUGAUGGUUAGGAAUUCAUUGGCCAUCGGCUAAGCAACAAUUUUUGGCAAUUGGAAAUCGA-GAGCGAAAGCUGAG-AUAUGCAAACUUUCUCUUUGA---GUGAAAGUCUCGAAAAA ......((((((((((..(((.((((((((((.......)))))))))).))).))).....))))))).........((((-((((....)))...-........((((((.(.....---).))))))))))).... ( -37.70, z-score = -2.18, R) >droYak2.chr3L 3170102 124 - 24197627 -----AGUAAAAUGACAGUGGGUGAAGGCUAGGAAUU----GGCUAUCGGCUGAUCCAAGUUUUUUGGCAAUUGGAAAUCGA-GAGCGAAAGCUAAG-AAAUGCAAACUUUCACUUUAA---GUGAAAGUCUAAAAAA- -----.(((......((((..(((((((((.(((.(.----.((.....))..)))).)))))))..)).))))....((..-.(((....)))..)-)..)))..((((((((.....---))))))))........- ( -33.80, z-score = -2.50, R) >droEre2.scaffold_4784 19311553 128 + 25762168 -----AGUAAAAUGACUGCGGGUGAAGGCUAGCCAUUCAUUGGCCAUCGGCUGAGCCACGUUUUUUGGCAAUUGGAAAUCGA-GAGCGAAAGCUAGG-AAAUGCAAACUUUCACUUUGA---GUGAAAGUCUAAAAAA- -----............((((((...((((.((((.....))))....))))..))).)))(((((((...(((.(..((..-.(((....)))..)-)..).)))((((((((.....---))))))))))))))).- ( -35.70, z-score = -0.97, R) >consensus ___________AUUACAGUGGGUGAUGGUUAGGAAUU_AUUGGCCAUCGGCUCAACAACAAUUUUUGGCAAUUGGAAAUCGA_GAGCGAAAGCUAAG_AAAUGAAAACUUUCACUUUGA___GUGAAAGUCUUGAAAA_ ....................................................................................(((....)))............((((((((........))))))))......... (-10.82 = -11.07 + 0.25)

| Location | 19,514,504 – 19,514,638 |
|---|---|
| Length | 134 |
| Sequences | 10 |
| Columns | 139 |
| Reading direction | reverse |
| Mean pairwise identity | 57.14 |
| Shannon entropy | 0.88445 |
| G+C content | 0.36982 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -8.24 |
| Energy contribution | -8.23 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.986951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
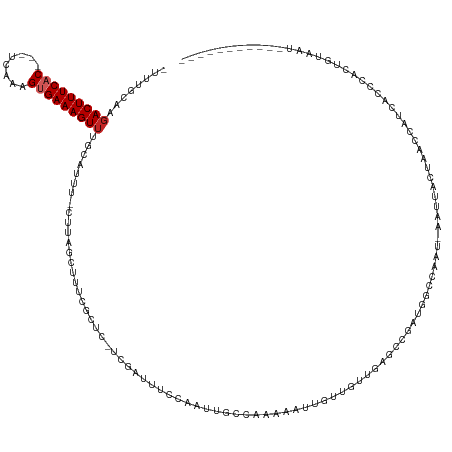
>dm3.chr3L 19514504 134 - 24543557 UUUUUCGAGACUUUCUC---UCAAAGUGAAAGUUUGCAUUU-CUUAGCUUUCGCUC-UCGAUUUCCAAUUGCCAAAAAUUGUUGCUUAUCCGAUGGCCACUAAAUUCCUAACCGUCACCCACAGCAAUUUUACUUAUUU ....((((((.......---.....(((((((((.......-...)))))))))))-)))).............((((((((((.......(((((...............))))).....))))))))))........ ( -25.46, z-score = -2.55, R) >droVir3.scaffold_13049 13089386 115 - 25233164 -UCUGCACUACUUUCACUUUUGGGGGUGAAAGUUUUUGGUC-GGCA-CUUUCGCUC-UUUAUCGC-----AGCCAAAAUUUGUUUUGUGUC---AAUGCGAUGGCAAAUAAAACGCAUUUGCCAAAA------------ -.(((.(((((((((((((....))))))))))....))))-))..-.........-...(((((-----((((((((....))))).)).---..))))))(((((((.......)))))))....------------ ( -29.90, z-score = -1.20, R) >droGri2.scaffold_15110 23826059 117 - 24565398 -UCUGCAUGACUUUCAC-UUUAGGGGUGAAAGUUUUUGCUA-AACA-CUUUCGCUC-CUAUUUAC-----GCCAAGUAUUGCUAUCGUGUUGUGGAUGCCUCGGCAAACAAAACACAAUUGACAAAU------------ -...(((..((((....-..((((((((((((((((....)-)).)-)))))))))-))).....-----...))))..)))....(((((.((..(((....)))..)).)))))...........------------ ( -34.24, z-score = -3.43, R) >droPer1.super_48 368972 124 - 581423 -UUUGCACGACUUUCAC--UCAAAAGUGAAAGUUUUCGUUUGCUUAACUUUCGCUUAACGAAAAAAAAAGGUCAACAAAUAUUAUUGU-UUAACAGCAAUGCAACCAUUCCUUAAAAUAUUGUUUAAC----------- -.(((((.(((((((((--(....)))))))))).....(((((....(((((.....)))))......(...(((((......))))-)...)))))))))))........................----------- ( -23.00, z-score = -1.94, R) >dp4.chrXR_group8 7590875 124 - 9212921 -UUUGCACGACUUUCAC--UCAAAAGUGAAAGUUUUCGUUUGCUUAACUUUCGCUUAACGAAAAAAAAAGGUCAACAAAUAUUAUUGU-UUAACAGCAAUGUAACCAUUCCUUAAAAUAUUGUUUAAC----------- -.(((.(((((((((((--(....))))))))((((((((.((.........))..))))))))...((((.......(((((.(((.-....))).))))).......))))......)))).))).----------- ( -22.14, z-score = -1.73, R) >droAna3.scaffold_13337 6434406 109 - 23293914 -UAUGCACGACUUUCAC---UCAAAGUGAAAGUUUUGUUUU-CCUAACUUUCGCUC-UCGAUUUCGAUUUGCCAAAAAUAUGUGUGCAACCAAUGAAUAA--AAUCUUGAAAACCCA---------------------- -..((((((((((((((---.....))))))))).......-..........((..-(((....)))...))...........)))))............--...............---------------------- ( -21.10, z-score = -2.48, R) >droSim1.chr3L 18844988 134 - 22553184 UUUUUCGAGACUUUCAC---UCAAAGUGAAAGUUUGCACAU-CUCAGCUUUCGCUC-UCGAUUUCCAAUUGCCAAAAAUUGUUGCUUAGCCGAUUGCCAAUAAAUUCCUAACCAUCACCCACUGCAAUUUUACUCAUUU ....(((((((((((((---.....))))))))........-...(((....))))-)))).....((((((.......((.((.((((..((((.......)))).)))).)).))......)))))).......... ( -22.52, z-score = -1.85, R) >droSec1.super_29 217596 134 - 700605 UUUUUCGAGACUUUCAC---UCAAAGAGAAAGUUUGCAUAU-CUCAGCUUUCGCUC-UCGAUUUCCAAUUGCCAAAAAUUGUUGCUUAGCCGAUGGCCAAUGAAUUCCUAACCAUCACCCACUGCAUUUUAACUCAUUU ......(((((((((.(---.....).)))))).((((...-....(((...((..-.((((((...........))))))..))..))).(((((...............)))))......))))......))).... ( -22.16, z-score = -0.54, R) >droYak2.chr3L 3170102 124 + 24197627 -UUUUUUAGACUUUCAC---UUAAAGUGAAAGUUUGCAUUU-CUUAGCUUUCGCUC-UCGAUUUCCAAUUGCCAAAAAACUUGGAUCAGCCGAUAGCC----AAUUCCUAGCCUUCACCCACUGUCAUUUUACU----- -.....(((((((((((---.....))))))))))).....-.((((..((.(((.-(((...(((((.(........).))))).....))).))).----))...)))).......................----- ( -21.70, z-score = -1.60, R) >droEre2.scaffold_4784 19311553 128 - 25762168 -UUUUUUAGACUUUCAC---UCAAAGUGAAAGUUUGCAUUU-CCUAGCUUUCGCUC-UCGAUUUCCAAUUGCCAAAAAACGUGGCUCAGCCGAUGGCCAAUGAAUGGCUAGCCUUCACCCGCAGUCAUUUUACU----- -.....(((((((((((---.....))))))))))).....-....(((.(((((.-..(.....)....((((.......))))..)).))).)))....((((((((.((........))))))))))....----- ( -29.20, z-score = -1.11, R) >consensus _UUUGCAAGACUUUCAC___UCAAAGUGAAAGUUUGCAUUU_CUUAGCUUUCGCUC_UCGAUUUCCAAUUGCCAAAAAUUGUUGUUGAGCCGAUGGCCAAU_AAUUACUAACCAUCACCCACUGUAAU___________ ........................((((((((((...........)))))))))).................................................................................... ( -8.24 = -8.23 + -0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:56 2011