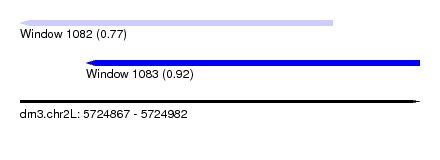
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,724,867 – 5,724,982 |
| Length | 115 |
| Max. P | 0.921715 |

| Location | 5,724,867 – 5,724,957 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 70.85 |
| Shannon entropy | 0.58982 |
| G+C content | 0.50603 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -10.98 |
| Energy contribution | -11.79 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.771991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
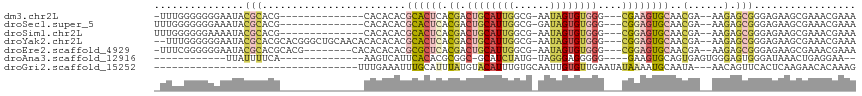
>dm3.chr2L 5724867 90 - 23011544 ACGCACUCACGACUGCAUUGGCGAAUAGUGUGGGCGAAGUGCAACGAAAGAGCGGGAGAAGCGAAACGAAAGACACGCGGUUUCAAUUUG ..(((((..((.(..(((((.....)))))..).)).)))))..(....)....((((..(((...(....)...)))..))))...... ( -25.60, z-score = -1.01, R) >droSec1.super_5 3803774 90 - 5866729 ACGCACUCACGACUGCAUUGGCGGAUAGUGUGGGCGGAGUGCAACGAAAGAGCGGGAGAAGCGAAACGAAAGACACGCGGUUUCAAUUUG ..((((((.((.(..(((((.....)))))..).))))))))..(....)....((((..(((...(....)...)))..))))...... ( -30.40, z-score = -2.62, R) >droSim1.chr2L 5520541 90 - 22036055 ACGCACUCACGACUGCAUUGGCGAAUAGUGUGGGCGGAGUGCAACGAAAGAGCGGGAGAAGCGAAACGAAAGACACGCGGUUUCAAUUUG ..((((((.((.(..(((((.....)))))..).))))))))..(....)....((((..(((...(....)...)))..))))...... ( -30.40, z-score = -2.65, R) >droYak2.chr2L 8857255 90 + 22324452 ACGCACUCACGACUGCAUUGGCGAAUAGUGUGGGCGGAGUGCAACGAAAGAGCGGGAGAAGCGAAACGAAAGACACGCGGUUUUAAUUUG ..((((((.((.(..(((((.....)))))..).))))))))..(....)....((((..(((...(....)...)))..))))...... ( -28.30, z-score = -2.31, R) >droEre2.scaffold_4929 5814864 90 - 26641161 ACGCGCUCACGACUGCAUUGGCGAAUAGUGUGGGCGGAGUGCAACGAAAGAGCGGGAGAAGCGAAACGAAAGACACGCGGUUUCAAUUUG ..((((((.((.(..(((((.....)))))..).))))))))..(....)....((((..(((...(....)...)))..))))...... ( -30.00, z-score = -1.93, R) >droAna3.scaffold_12916 11167588 84 - 16180835 AUUCACACGCGGC-GCAUCUAUGUAGGGAGGGGG-GAAGUGCAGUGAGUGGGAGUGGGA--UAAACUGAGGAAGACACUUUUCUAUUG-- .(((((.(((.((-((.(((.............)-)).)))).))).)))))(((((((--......(((.......)))))))))).-- ( -17.52, z-score = 0.32, R) >droGri2.scaffold_15252 3267630 77 - 17193109 ------ACAUUUGUGCAAUUGUGUUGAAUAU----AAAAUGCAAUAAACAGUUCACUCAAG-AACACAAAGGAAACGCGUUUGCUGCG-- ------........(((((((((((......----..))))))))((((.((((......)-))).....(....)..))))..))).-- ( -14.90, z-score = -0.08, R) >consensus ACGCACUCACGACUGCAUUGGCGAAUAGUGUGGGCGGAGUGCAACGAAAGAGCGGGAGAAGCGAAACGAAAGACACGCGGUUUCAAUUUG ..((((...((.((((((((.....)))))))).))..)))).............((((.(((...(....)...)))..))))...... (-10.98 = -11.79 + 0.80)

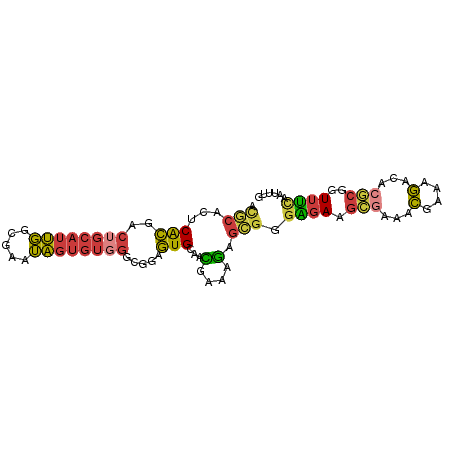
| Location | 5,724,886 – 5,724,982 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 62.50 |
| Shannon entropy | 0.66531 |
| G+C content | 0.51167 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -10.63 |
| Energy contribution | -11.07 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.921715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5724886 96 - 23011544 -UUUGGGGGGGAAUACGCACG--------------CACACACGCACUCACGACUGCAUUGGCG-AAUAGUGUGGG---CGAAGUGCAACGA--AAGAGCGGGAGAAGCGAAACGAAA -..............(((.((--------------(......(((((..((.(..(((((...-..)))))..).---)).)))))..(..--..).)))......)))........ ( -23.80, z-score = -0.57, R) >droSec1.super_5 3803793 97 - 5866729 UUUGGGGGGGAAAUACGCACG--------------CACACACGCACUCACGACUGCAUUGGCG-GAUAGUGUGGG---CGGAGUGCAACGA--AAGAGCGGGAGAAGCGAAACGAAA ...............(((.((--------------(......((((((.((.(..(((((...-..)))))..).---))))))))..(..--..).)))......)))........ ( -28.60, z-score = -2.29, R) >droSim1.chr2L 5520560 97 - 22036055 UUUGGGGGGAAAAUACGCACG--------------CACACACGCACUCACGACUGCAUUGGCG-AAUAGUGUGGG---CGGAGUGCAACGA--AAGAGCGGGAGAAGCGAAACGAAA ...............(((.((--------------(......((((((.((.(..(((((...-..)))))..).---))))))))..(..--..).)))......)))........ ( -28.60, z-score = -2.40, R) >droYak2.chr2L 8857274 109 + 22324452 --UUUGGGGGGAAUACGCACGCACGGGCUGCAACACACACACGCACUCACGACUGCAUUGGCG-AAUAGUGUGGG---CGGAGUGCAACGA--AAGAGCGGGAGAAGCGAAACGAAA --(((((.(......).).(((.....((((...........((((((.((.(..(((((...-..)))))..).---))))))))..(..--..).)))).....)))...)))). ( -32.30, z-score = -1.89, R) >droEre2.scaffold_4929 5814883 102 - 26641161 -UUUCGGGGGGAAUACGCACGCACG--------CACACACACGCGCUCACGACUGCAUUGGCG-AAUAGUGUGGG---CGGAGUGCAACGA--AAGAGCGGGAGAAGCGAAACGAAA -.(((((.(......).).(((.((--------(........((((((.((.(..(((((...-..)))))..).---))))))))..(..--..).)))......)))...)))). ( -32.00, z-score = -1.73, R) >droAna3.scaffold_12916 11167603 83 - 16180835 ------------UUAUUUUCA--------------AAGUCAUUCACACGCGGC-GCAUCUAUG-UAGGGAGGGGG----GAAGUGCAGUGAGUGGGAGUGGGAUAAACUGAGGAA-- ------------....(..((--------------...((((((((.(((..(-.(.(((...-...)))...).----)..)))..))))))))...))..)............-- ( -18.40, z-score = -0.98, R) >droGri2.scaffold_15252 3267647 80 - 17193109 ----------------------------------UUUGAAAUUUGCAUUUAUGUACAUUUGUGCAAUUGUGUUGAAUAUAAAAUGCAAUA---AACAGUUCACUCAAGAACACAAAG ----------------------------------((((....((((((((.(((((....)))))..(((((...)))))))))))))..---....((((......)))).)))). ( -16.00, z-score = -1.10, R) >consensus _UUUGGGGGGGAAUACGCACG______________CACACACGCACUCACGACUGCAUUGGCG_AAUAGUGUGGG___CGGAGUGCAACGA__AAGAGCGGGAGAAGCGAAACGAAA ..........................................((((((.((.((((((((......))))))))....))))))))...........((.......))......... (-10.63 = -11.07 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:45 2011