
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,482,488 – 19,482,632 |
| Length | 144 |
| Max. P | 0.956775 |

| Location | 19,482,488 – 19,482,632 |
|---|---|
| Length | 144 |
| Sequences | 5 |
| Columns | 144 |
| Reading direction | forward |
| Mean pairwise identity | 68.92 |
| Shannon entropy | 0.57126 |
| G+C content | 0.60718 |
| Mean single sequence MFE | -51.98 |
| Consensus MFE | -22.21 |
| Energy contribution | -23.69 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
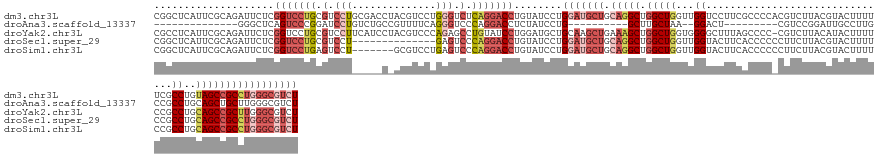
>dm3.chr3L 19482488 144 + 24543557 AGACGCCCAGGCGGCUACAGGCGAAAAAGUACGUAAGACGUGGGGCGAAGGACCAACCAGCCAGCCUGCAGCAUCCAGGAUACAGGUCCUGAGACCCAGGACGUAGGUCGCAGGACGCAGGACCGAGAAUCUGCGAAUGAGCCG ...(((....)))(((.(((((.......((((.....)))).(((...((.....)).))).))))).))).....((...((.((((((.((((.........)))).))))))(((((........)))))...))..)). ( -51.00, z-score = -1.37, R) >droAna3.scaffold_13337 5291826 109 - 23293914 AGACGCCCAAGCAGCUGCAGGCGGCAAGGCAAUCCGGACG---------AGUCC--UUAGCAAGC----------CAGGAUAGAGGUCCUGGGACCCUGAAAACGGCAGACAGGAUCCGGGACUGAGCCC-------------- ...((((...((....)).))))....(((.((((((.(.---------.((..--...))..))----------).))))...(((((((((..((((...........)))).)))))))))..))).-------------- ( -36.20, z-score = 0.30, R) >droYak2.chr3L 3140702 143 - 24197627 AGACGCCCAAGCGGCUGCAGGCGGAAAAGUAUGUAAGACG-GGGGCUAAAGCCCCACCAGCCAGCUUUCAGCUUGCAGCAUCCAGGAUACAGGCUCUGGGACGUAGGAUGAAGGACGCAGGACCGAGAAUCUGCGAAUGAGGCG ...((((((.((((((((((((.((((.((.((.......-(((((....)))))......)))))))).))))))))).((((((........)))))).)))...........((((((........))))))..)).)))) ( -54.72, z-score = -1.73, R) >droSec1.super_29 186851 130 + 700605 AGACGCCCAGGCGGCUGCAGGCGGAAAAGUACGUAAGAAGGGGGGUGAAGUACCAACCAGCCAGCCUGCAGCAUCCAGGAUACAGGUCCUGGGACUC--------------AGGACGCAGGACCGAGAAUCUGCGAAUGAGCCG ...(((....)))(((((((((((.......(....)..((..((((...))))..))..)).))))))))).((((((((....)))))))).(((--------------(...((((((........))))))..))))... ( -57.40, z-score = -3.76, R) >droSim1.chr3L 18814757 137 + 22553184 AGACGCCCAGGCGGCUGCAGGCGGAAAAGUACGUAAGAAGGGGGGUGAAGUACCAACCAGCCAGCCUGCAGCAUCCAGGAUACAGGUCCUGGGACUCAGGACGC-------AGGACUCAGGACCGAGAAUCUGCGAAUGAGCCG ...(((....)))(((((((((((.......(....)..((..((((...))))..))..)).))))))))).((((((((....)))))))).((((...(((-------(((.(((......)))..))))))..))))... ( -60.60, z-score = -3.76, R) >consensus AGACGCCCAGGCGGCUGCAGGCGGAAAAGUACGUAAGACG_GGGGCGAAGGACCAACCAGCCAGCCUGCAGCAUCCAGGAUACAGGUCCUGGGACCCAGGACGC_G_____AGGACGCAGGACCGAGAAUCUGCGAAUGAGCCG ...(((.((((((((((...((......))............((........))...))))).)))))................(((((((.(.((................)).).)))))))........)))......... (-22.21 = -23.69 + 1.48)


| Location | 19,482,488 – 19,482,632 |
|---|---|
| Length | 144 |
| Sequences | 5 |
| Columns | 144 |
| Reading direction | reverse |
| Mean pairwise identity | 68.92 |
| Shannon entropy | 0.57126 |
| G+C content | 0.60718 |
| Mean single sequence MFE | -50.07 |
| Consensus MFE | -21.18 |
| Energy contribution | -24.98 |
| Covariance contribution | 3.80 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19482488 144 - 24543557 CGGCUCAUUCGCAGAUUCUCGGUCCUGCGUCCUGCGACCUACGUCCUGGGUCUCAGGACCUGUAUCCUGGAUGCUGCAGGCUGGCUGGUUGGUCCUUCGCCCCACGUCUUACGUACUUUUUCGCCUGUAGCCGCCUGGGCGUCU ..(((((...((((..(....)..))))((((((.((((((.....)))))).)))))).........((..(((((((((.(((.((.....))...)))..(((.....)))........)))))))))..))))))).... ( -56.20, z-score = -1.88, R) >droAna3.scaffold_13337 5291826 109 + 23293914 --------------GGGCUCAGUCCCGGAUCCUGUCUGCCGUUUUCAGGGUCCCAGGACCUCUAUCCUG----------GCUUGCUAA--GGACU---------CGUCCGGAUUGCCUUGCCGCCUGCAGCUGCUUGGGCGUCU --------------((((..(((((.((((((((...........))))))))(((((......)))))----------.........--)))))---------.))))((((.((((.((.((.....)).))..)))))))) ( -42.90, z-score = -0.78, R) >droYak2.chr3L 3140702 143 + 24197627 CGCCUCAUUCGCAGAUUCUCGGUCCUGCGUCCUUCAUCCUACGUCCCAGAGCCUGUAUCCUGGAUGCUGCAAGCUGAAAGCUGGCUGGUGGGGCUUUAGCCCC-CGUCUUACAUACUUUUCCGCCUGCAGCCGCUUGGGCGUCU ((((.((..(((((..(((.((....((((..........)))))).)))..)))).........((((((.((.(((((.((...((.(((((....)))))-...))....)).))))).)).)))))).)..))))))... ( -45.00, z-score = 0.30, R) >droSec1.super_29 186851 130 - 700605 CGGCUCAUUCGCAGAUUCUCGGUCCUGCGUCCU--------------GAGUCCCAGGACCUGUAUCCUGGAUGCUGCAGGCUGGCUGGUUGGUACUUCACCCCCCUUCUUACGUACUUUUCCGCCUGCAGCCGCCUGGGCGUCU .((((((..(((((..(....)..)))))...)--------------)))))((((((......))))))..(((((((((.((......(((((.................)))))...)))))))))))(((....)))... ( -50.93, z-score = -2.29, R) >droSim1.chr3L 18814757 137 - 22553184 CGGCUCAUUCGCAGAUUCUCGGUCCUGAGUCCU-------GCGUCCUGAGUCCCAGGACCUGUAUCCUGGAUGCUGCAGGCUGGCUGGUUGGUACUUCACCCCCCUUCUUACGUACUUUUCCGCCUGCAGCCGCCUGGGCGUCU .((((((..(((((...((((....))))..))-------)))...))))))((((((......))))))..(((((((((.((......(((((.................)))))...)))))))))))(((....)))... ( -55.33, z-score = -2.71, R) >consensus CGGCUCAUUCGCAGAUUCUCGGUCCUGCGUCCU_____C_ACGUCCUGGGUCCCAGGACCUGUAUCCUGGAUGCUGCAGGCUGGCUGGUUGGUCCUUCACCCC_CGUCUUACGUACUUUUCCGCCUGCAGCCGCCUGGGCGUCU ....................(((((((.(.(((..............))).).)))))))........(((((((.(((((.(((((...((...............................))..))))))))))))))))) (-21.18 = -24.98 + 3.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:52 2011